| Name | cluster:GABPA.H14CORE.0.PSM.A |
| Cluster logo | 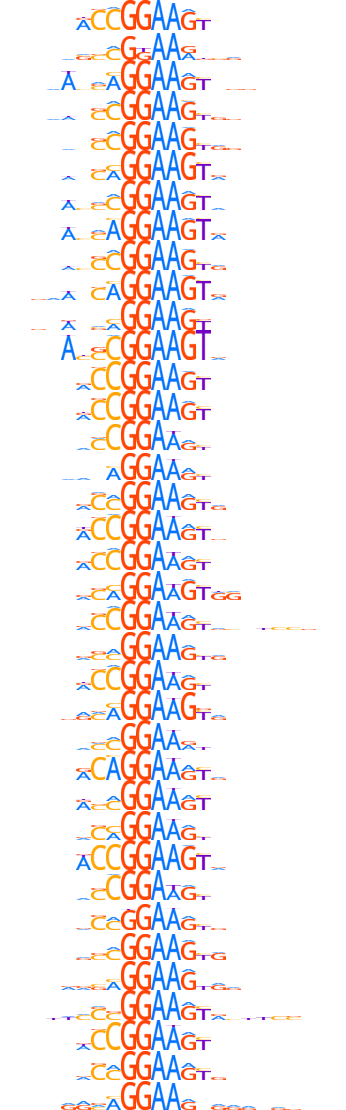 |
| Representative motif logo |  |
| Representative motif | GABPA.H14CORE.0.PSM.A |
| Representative motif gene symbol | GABPA |
| Clustered motifs | E4F1.H14CORE.0.P.B,
EHF.H14CORE.0.P.B,
ELF1.H14CORE.0.PSM.A,
ELF2.H14CORE.0.PS.A,
ELF2.H14CORE.1.M.B,
ELF3.H14CORE.0.S.B,
ELF3.H14CORE.1.PM.A,
ELF4.H14CORE.0.PS.A,
ELF4.H14CORE.1.M.B,
ELF5.H14CORE.0.PSM.A,
ELF5.H14CORE.1.S.B,
ELK1.H14CORE.0.PSM.A,
ELK3.H14CORE.0.PSM.A,
ELK4.H14CORE.0.PSM.A,
ERF.H14CORE.0.PS.A,
ERG.H14CORE.0.P.B,
ERG.H14CORE.1.SM.B,
ETS1.H14CORE.0.S.B,
ETS1.H14CORE.1.P.B,
ETS2.H14CORE.0.S.C,
ETS2.H14CORE.1.P.B,
ETV1.H14CORE.0.PSM.A,
ETV1.H14CORE.1.PM.A,
ETV2.H14CORE.0.S.B,
ETV2.H14CORE.1.PM.A,
ETV3.H14CORE.0.SM.B,
ETV4.H14CORE.0.P.B,
ETV4.H14CORE.1.SM.B,
ETV5.H14CORE.0.PS.A,
ETV5.H14CORE.1.PM.A,
ETV6.H14CORE.0.PS.A,
ETV6.H14CORE.1.P.B,
ETV7.H14CORE.0.SM.B,
FEV.H14CORE.0.S.B,
FLI1.H14CORE.0.PSM.A,
FLI1.H14CORE.1.P.B,
GABPA.H14CORE.0.PSM.A |
| Clustered motifs gene symbols plus families | E4F1 {2.3.4.0.74} (TFClass),
ETS1 {3.5.2.1.1} (TFClass),
ERF {3.5.2.1.11} (TFClass),
ETS2 {3.5.2.1.2} (TFClass),
ETV2 {3.5.2.1.4} (TFClass),
GABPA {3.5.2.1.5} (TFClass),
FLI1 {3.5.2.1.6} (TFClass),
ERG {3.5.2.1.7} (TFClass),
FEV {3.5.2.1.8} (TFClass),
ETV3 {3.5.2.1.9} (TFClass),
ELK1 {3.5.2.2.1} (TFClass),
ELK3 {3.5.2.2.2} (TFClass),
ELK4 {3.5.2.2.3} (TFClass),
ETV1 {3.5.2.2.4} (TFClass),
ETV4 {3.5.2.2.5} (TFClass),
ETV5 {3.5.2.2.6} (TFClass),
ELF1 {3.5.2.3.1} (TFClass),
ELF2 {3.5.2.3.2} (TFClass),
ELF4 {3.5.2.3.3} (TFClass),
EHF {3.5.2.4.1} (TFClass),
ELF3 {3.5.2.4.2} (TFClass),
ELF5 {3.5.2.4.3} (TFClass),
ETV6 {3.5.2.6.1} (TFClass),
ETV7 {3.5.2.6.2} (TFClass) |
| Primary family | Ets-related {3.5.2} (TFClass) |
| Primary subfamily | ETS-like {3.5.2.1} (TFClass) |
| Cluster size | 37 |
| Average similarity | 0.444 |
| Min similarity | 0.207 |