| Motif | ZSCA2.H14INVIVO.0.PG.A |
| Gene (human) | ZSCAN2 (GeneCards) |
| Gene synonyms (human) | ZFP29, ZNF854 |
| Gene (mouse) | Zscan2 |
| Gene synonyms (mouse) | Zfp-29, Zfp29 |
| LOGO |  |
| LOGO (reverse complement) | 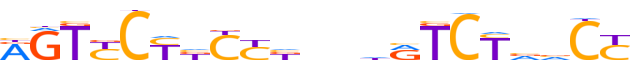 |
| Motif subtype | 0 |
| Quality | A |
| Motif | ZSCA2.H14INVIVO.0.PG.A |
| Gene (human) | ZSCAN2 (GeneCards) |
| Gene synonyms (human) | ZFP29, ZNF854 |
| Gene (mouse) | Zscan2 |
| Gene synonyms (mouse) | Zfp-29, Zfp29 |
| LOGO |  |
| LOGO (reverse complement) | 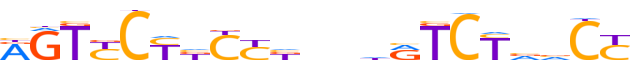 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 21 |
| Consensus | RGbbRGAYhnnvRGvRGRACY |
| GC content | 51.77% |
| Information content (bits; total / per base) | 16.091 / 0.766 |
| Data sources | ChIP-Seq + Genomic HT-SELEX |
| Aligned words | 500 |
| Previous names |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|
| Overall | 3 (3) | 0.77 | 0.871 | 0.714 | 0.857 | 0.707 | 0.9 | 23.886 | 104.854 |
| Genomic HT-SELEX benchmarking | Centrality | pseudo-auROC | auROC | auPR | |
|---|---|---|---|---|---|
| Lysate, 2 experiments | median | 112.063 | 0.782 | 0.774 | 0.793 |
| best | 176.959 | 0.932 | 0.92 | 0.902 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZNF180-like {2.3.3.58} (TFClass) |
| TFClass ID | TFClass: 2.3.3.58.6 |
| HGNC | HGNC:20994 |
| MGI | MGI:99176 |
| EntrezGene (human) | GeneID:54993 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:22691 (SSTAR profile) |
| UniProt ID (human) | ZSCA2_HUMAN |
| UniProt ID (mouse) | ZSCA2_MOUSE |
| UniProt AC (human) | Q7Z7L9 (TFClass) |
| UniProt AC (mouse) | Q07230 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 3 human, 0 mouse |
| HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| Genomic HT-SELEX | 2 overall: 2 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | ZSCA2.H14INVIVO.0.PG.A.pcm |
| PWM | ZSCA2.H14INVIVO.0.PG.A.pwm |
| PFM | ZSCA2.H14INVIVO.0.PG.A.pfm |
| Threshold to P-value map | ZSCA2.H14INVIVO.0.PG.A.thr |
| Motif in other formats | |
| JASPAR format | ZSCA2.H14INVIVO.0.PG.A_jaspar_format.txt |
| MEME format | ZSCA2.H14INVIVO.0.PG.A_meme_format.meme |
| Transfac format | ZSCA2.H14INVIVO.0.PG.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 193.0 | 3.0 | 291.0 | 13.0 |
| 02 | 23.0 | 9.0 | 454.0 | 14.0 |
| 03 | 60.0 | 97.0 | 104.0 | 239.0 |
| 04 | 40.0 | 131.0 | 78.0 | 251.0 |
| 05 | 382.0 | 6.0 | 75.0 | 37.0 |
| 06 | 9.0 | 4.0 | 472.0 | 15.0 |
| 07 | 438.0 | 33.0 | 21.0 | 8.0 |
| 08 | 17.0 | 302.0 | 47.0 | 134.0 |
| 09 | 205.0 | 104.0 | 57.0 | 134.0 |
| 10 | 118.0 | 113.0 | 117.0 | 152.0 |
| 11 | 115.0 | 183.0 | 133.0 | 69.0 |
| 12 | 284.0 | 82.0 | 78.0 | 56.0 |
| 13 | 159.0 | 13.0 | 305.0 | 23.0 |
| 14 | 65.0 | 23.0 | 402.0 | 10.0 |
| 15 | 289.0 | 92.0 | 69.0 | 50.0 |
| 16 | 323.0 | 3.0 | 141.0 | 33.0 |
| 17 | 12.0 | 2.0 | 480.0 | 6.0 |
| 18 | 179.0 | 40.0 | 278.0 | 3.0 |
| 19 | 436.0 | 39.0 | 20.0 | 5.0 |
| 20 | 15.0 | 420.0 | 19.0 | 46.0 |
| 21 | 43.0 | 56.0 | 40.0 | 361.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.386 | 0.006 | 0.582 | 0.026 |
| 02 | 0.046 | 0.018 | 0.908 | 0.028 |
| 03 | 0.12 | 0.194 | 0.208 | 0.478 |
| 04 | 0.08 | 0.262 | 0.156 | 0.502 |
| 05 | 0.764 | 0.012 | 0.15 | 0.074 |
| 06 | 0.018 | 0.008 | 0.944 | 0.03 |
| 07 | 0.876 | 0.066 | 0.042 | 0.016 |
| 08 | 0.034 | 0.604 | 0.094 | 0.268 |
| 09 | 0.41 | 0.208 | 0.114 | 0.268 |
| 10 | 0.236 | 0.226 | 0.234 | 0.304 |
| 11 | 0.23 | 0.366 | 0.266 | 0.138 |
| 12 | 0.568 | 0.164 | 0.156 | 0.112 |
| 13 | 0.318 | 0.026 | 0.61 | 0.046 |
| 14 | 0.13 | 0.046 | 0.804 | 0.02 |
| 15 | 0.578 | 0.184 | 0.138 | 0.1 |
| 16 | 0.646 | 0.006 | 0.282 | 0.066 |
| 17 | 0.024 | 0.004 | 0.96 | 0.012 |
| 18 | 0.358 | 0.08 | 0.556 | 0.006 |
| 19 | 0.872 | 0.078 | 0.04 | 0.01 |
| 20 | 0.03 | 0.84 | 0.038 | 0.092 |
| 21 | 0.086 | 0.112 | 0.08 | 0.722 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.43 | -3.325 | 0.838 | -2.163 |
| 02 | -1.64 | -2.484 | 1.281 | -2.096 |
| 03 | -0.721 | -0.25 | -0.181 | 0.642 |
| 04 | -1.114 | 0.046 | -0.464 | 0.691 |
| 05 | 1.109 | -2.819 | -0.503 | -1.189 |
| 06 | -2.484 | -3.126 | 1.32 | -2.034 |
| 07 | 1.245 | -1.298 | -1.725 | -2.584 |
| 08 | -1.92 | 0.875 | -0.958 | 0.069 |
| 09 | 0.49 | -0.181 | -0.771 | 0.069 |
| 10 | -0.057 | -0.1 | -0.065 | 0.193 |
| 11 | -0.082 | 0.377 | 0.061 | -0.584 |
| 12 | 0.814 | -0.415 | -0.464 | -0.788 |
| 13 | 0.238 | -2.163 | 0.885 | -1.64 |
| 14 | -0.643 | -1.64 | 1.16 | -2.394 |
| 15 | 0.831 | -0.302 | -0.584 | -0.898 |
| 16 | 0.942 | -3.325 | 0.119 | -1.298 |
| 17 | -2.234 | -3.573 | 1.336 | -2.819 |
| 18 | 0.355 | -1.114 | 0.793 | -3.325 |
| 19 | 1.241 | -1.138 | -1.77 | -2.961 |
| 20 | -2.034 | 1.203 | -1.818 | -0.979 |
| 21 | -1.044 | -0.788 | -1.114 | 1.053 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.96081 |
| 0.0005 | 4.06506 |
| 0.0001 | 6.39961 |