| Motif | ZN708.H14CORE.0.P.B |
| Gene (human) | ZNF708 (GeneCards) |
| Gene synonyms (human) | KOX8, ZNF15, ZNF15L1 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 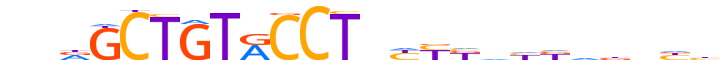 |
| Motif subtype | 0 |
| Quality | B |
| Motif | ZN708.H14CORE.0.P.B |
| Gene (human) | ZNF708 (GeneCards) |
| Gene synonyms (human) | KOX8, ZNF15, ZNF15L1 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 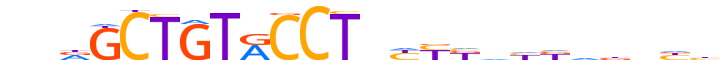 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 24 |
| Consensus | vvnvhvddvRRnAGGYACAGChnn |
| GC content | 51.77% |
| Information content (bits; total / per base) | 16.921 / 0.705 |
| Data sources | ChIP-Seq |
| Aligned words | 780 |
| Previous names | ZN708.H12CORE.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (9) | 0.831 | 0.913 | 0.839 | 0.902 | 0.822 | 0.92 | 6.557 | 7.35 | 363.77 | 396.125 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.181 |
| HGNC | HGNC:12945 |
| MGI | |
| EntrezGene (human) | GeneID:7562 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN708_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | P17019 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | ZN708.H14CORE.0.P.B.pcm |
| PWM | ZN708.H14CORE.0.P.B.pwm |
| PFM | ZN708.H14CORE.0.P.B.pfm |
| Threshold to P-value map | ZN708.H14CORE.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | ZN708.H14CORE.0.P.B_jaspar_format.txt |
| MEME format | ZN708.H14CORE.0.P.B_meme_format.meme |
| Transfac format | ZN708.H14CORE.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 210.0 | 117.0 | 341.0 | 112.0 |
| 02 | 146.0 | 101.0 | 434.0 | 99.0 |
| 03 | 178.0 | 298.0 | 180.0 | 124.0 |
| 04 | 183.0 | 355.0 | 124.0 | 118.0 |
| 05 | 146.0 | 149.0 | 119.0 | 366.0 |
| 06 | 442.0 | 121.0 | 109.0 | 108.0 |
| 07 | 396.0 | 69.0 | 212.0 | 103.0 |
| 08 | 145.0 | 125.0 | 180.0 | 330.0 |
| 09 | 469.0 | 120.0 | 102.0 | 89.0 |
| 10 | 467.0 | 60.0 | 216.0 | 37.0 |
| 11 | 198.0 | 35.0 | 437.0 | 110.0 |
| 12 | 117.0 | 219.0 | 216.0 | 228.0 |
| 13 | 733.0 | 2.0 | 40.0 | 5.0 |
| 14 | 8.0 | 1.0 | 764.0 | 7.0 |
| 15 | 14.0 | 28.0 | 732.0 | 6.0 |
| 16 | 10.0 | 323.0 | 5.0 | 442.0 |
| 17 | 742.0 | 11.0 | 13.0 | 14.0 |
| 18 | 4.0 | 682.0 | 13.0 | 81.0 |
| 19 | 721.0 | 2.0 | 50.0 | 7.0 |
| 20 | 26.0 | 4.0 | 742.0 | 8.0 |
| 21 | 44.0 | 675.0 | 21.0 | 40.0 |
| 22 | 156.0 | 230.0 | 20.0 | 374.0 |
| 23 | 191.0 | 154.0 | 305.0 | 130.0 |
| 24 | 193.0 | 136.0 | 300.0 | 151.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.269 | 0.15 | 0.437 | 0.144 |
| 02 | 0.187 | 0.129 | 0.556 | 0.127 |
| 03 | 0.228 | 0.382 | 0.231 | 0.159 |
| 04 | 0.235 | 0.455 | 0.159 | 0.151 |
| 05 | 0.187 | 0.191 | 0.153 | 0.469 |
| 06 | 0.567 | 0.155 | 0.14 | 0.138 |
| 07 | 0.508 | 0.088 | 0.272 | 0.132 |
| 08 | 0.186 | 0.16 | 0.231 | 0.423 |
| 09 | 0.601 | 0.154 | 0.131 | 0.114 |
| 10 | 0.599 | 0.077 | 0.277 | 0.047 |
| 11 | 0.254 | 0.045 | 0.56 | 0.141 |
| 12 | 0.15 | 0.281 | 0.277 | 0.292 |
| 13 | 0.94 | 0.003 | 0.051 | 0.006 |
| 14 | 0.01 | 0.001 | 0.979 | 0.009 |
| 15 | 0.018 | 0.036 | 0.938 | 0.008 |
| 16 | 0.013 | 0.414 | 0.006 | 0.567 |
| 17 | 0.951 | 0.014 | 0.017 | 0.018 |
| 18 | 0.005 | 0.874 | 0.017 | 0.104 |
| 19 | 0.924 | 0.003 | 0.064 | 0.009 |
| 20 | 0.033 | 0.005 | 0.951 | 0.01 |
| 21 | 0.056 | 0.865 | 0.027 | 0.051 |
| 22 | 0.2 | 0.295 | 0.026 | 0.479 |
| 23 | 0.245 | 0.197 | 0.391 | 0.167 |
| 24 | 0.247 | 0.174 | 0.385 | 0.194 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.074 | -0.505 | 0.555 | -0.548 |
| 02 | -0.287 | -0.65 | 0.795 | -0.67 |
| 03 | -0.09 | 0.421 | -0.079 | -0.448 |
| 04 | -0.063 | 0.595 | -0.448 | -0.497 |
| 05 | -0.287 | -0.266 | -0.488 | 0.626 |
| 06 | 0.814 | -0.472 | -0.575 | -0.584 |
| 07 | 0.704 | -1.024 | 0.083 | -0.631 |
| 08 | -0.293 | -0.44 | -0.079 | 0.523 |
| 09 | 0.873 | -0.48 | -0.64 | -0.774 |
| 10 | 0.868 | -1.16 | 0.101 | -1.627 |
| 11 | 0.015 | -1.68 | 0.802 | -0.566 |
| 12 | -0.505 | 0.115 | 0.101 | 0.155 |
| 13 | 1.318 | -3.983 | -1.552 | -3.385 |
| 14 | -3.013 | -4.301 | 1.359 | -3.122 |
| 15 | -2.53 | -1.892 | 1.317 | -3.245 |
| 16 | -2.825 | 0.501 | -3.385 | 0.814 |
| 17 | 1.33 | -2.743 | -2.596 | -2.53 |
| 18 | -3.547 | 1.246 | -2.596 | -0.867 |
| 19 | 1.301 | -3.983 | -1.337 | -3.122 |
| 20 | -1.961 | -3.547 | 1.33 | -3.013 |
| 21 | -1.46 | 1.236 | -2.161 | -1.552 |
| 22 | -0.221 | 0.164 | -2.206 | 0.647 |
| 23 | -0.021 | -0.234 | 0.444 | -0.401 |
| 24 | -0.01 | -0.357 | 0.428 | -0.253 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.27306 |
| 0.0005 | 3.49506 |
| 0.0001 | 6.06941 |