| Motif | ZN547.H14CORE.0.P.B |
| Gene (human) | ZNF547 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 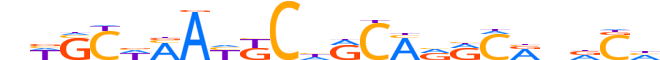 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | ZN547.H14CORE.0.P.B |
| Gene (human) | ZNF547 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 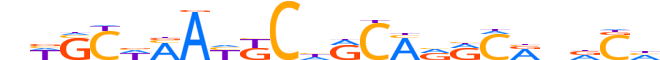 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 22 |
| Consensus | nWSCWMAYKChRCMRRCRndCh |
| GC content | 51.09% |
| Information content (bits; total / per base) | 16.464 / 0.748 |
| Data sources | ChIP-Seq |
| Aligned words | 731 |
| Previous names | ZN547.H12CORE.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 3 (15) | 0.815 | 0.895 | 0.791 | 0.85 | 0.785 | 0.853 | 5.354 | 5.848 | 260.699 | 355.886 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | Unclassified {2.3.3.0} (TFClass) |
| TFClass ID | TFClass: 2.3.3.0.97 |
| HGNC | HGNC:26432 |
| MGI | |
| EntrezGene (human) | GeneID:284306 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZN547_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q8IVP9 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 3 human, 0 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | ZN547.H14CORE.0.P.B.pcm |
| PWM | ZN547.H14CORE.0.P.B.pwm |
| PFM | ZN547.H14CORE.0.P.B.pfm |
| Threshold to P-value map | ZN547.H14CORE.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | ZN547.H14CORE.0.P.B_jaspar_format.txt |
| MEME format | ZN547.H14CORE.0.P.B_meme_format.meme |
| Transfac format | ZN547.H14CORE.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 105.0 | 155.0 | 251.0 | 220.0 |
| 02 | 99.0 | 86.0 | 73.0 | 473.0 |
| 03 | 50.0 | 72.0 | 576.0 | 33.0 |
| 04 | 24.0 | 596.0 | 2.0 | 109.0 |
| 05 | 200.0 | 64.0 | 58.0 | 409.0 |
| 06 | 533.0 | 89.0 | 75.0 | 34.0 |
| 07 | 695.0 | 6.0 | 28.0 | 2.0 |
| 08 | 105.0 | 144.0 | 27.0 | 455.0 |
| 09 | 3.0 | 38.0 | 402.0 | 288.0 |
| 10 | 2.0 | 728.0 | 1.0 | 0.0 |
| 11 | 283.0 | 118.0 | 53.0 | 277.0 |
| 12 | 178.0 | 23.0 | 492.0 | 38.0 |
| 13 | 10.0 | 650.0 | 14.0 | 57.0 |
| 14 | 572.0 | 82.0 | 20.0 | 57.0 |
| 15 | 237.0 | 63.0 | 386.0 | 45.0 |
| 16 | 159.0 | 46.0 | 475.0 | 51.0 |
| 17 | 75.0 | 596.0 | 19.0 | 41.0 |
| 18 | 481.0 | 58.0 | 135.0 | 57.0 |
| 19 | 132.0 | 168.0 | 157.0 | 274.0 |
| 20 | 341.0 | 48.0 | 272.0 | 70.0 |
| 21 | 30.0 | 591.0 | 63.0 | 47.0 |
| 22 | 386.0 | 164.0 | 52.0 | 129.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.144 | 0.212 | 0.343 | 0.301 |
| 02 | 0.135 | 0.118 | 0.1 | 0.647 |
| 03 | 0.068 | 0.098 | 0.788 | 0.045 |
| 04 | 0.033 | 0.815 | 0.003 | 0.149 |
| 05 | 0.274 | 0.088 | 0.079 | 0.56 |
| 06 | 0.729 | 0.122 | 0.103 | 0.047 |
| 07 | 0.951 | 0.008 | 0.038 | 0.003 |
| 08 | 0.144 | 0.197 | 0.037 | 0.622 |
| 09 | 0.004 | 0.052 | 0.55 | 0.394 |
| 10 | 0.003 | 0.996 | 0.001 | 0.0 |
| 11 | 0.387 | 0.161 | 0.073 | 0.379 |
| 12 | 0.244 | 0.031 | 0.673 | 0.052 |
| 13 | 0.014 | 0.889 | 0.019 | 0.078 |
| 14 | 0.782 | 0.112 | 0.027 | 0.078 |
| 15 | 0.324 | 0.086 | 0.528 | 0.062 |
| 16 | 0.218 | 0.063 | 0.65 | 0.07 |
| 17 | 0.103 | 0.815 | 0.026 | 0.056 |
| 18 | 0.658 | 0.079 | 0.185 | 0.078 |
| 19 | 0.181 | 0.23 | 0.215 | 0.375 |
| 20 | 0.466 | 0.066 | 0.372 | 0.096 |
| 21 | 0.041 | 0.808 | 0.086 | 0.064 |
| 22 | 0.528 | 0.224 | 0.071 | 0.176 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.548 | -0.163 | 0.315 | 0.184 |
| 02 | -0.605 | -0.744 | -0.904 | 0.945 |
| 03 | -1.273 | -0.918 | 1.142 | -1.672 |
| 04 | -1.973 | 1.176 | -3.923 | -0.511 |
| 05 | 0.089 | -1.033 | -1.129 | 0.801 |
| 06 | 1.065 | -0.71 | -0.878 | -1.643 |
| 07 | 1.329 | -3.183 | -1.828 | -3.923 |
| 08 | -0.548 | -0.236 | -1.862 | 0.907 |
| 09 | -3.681 | -1.537 | 0.783 | 0.452 |
| 10 | -3.923 | 1.375 | -4.243 | -4.717 |
| 11 | 0.434 | -0.433 | -1.216 | 0.413 |
| 12 | -0.026 | -2.012 | 0.985 | -1.537 |
| 13 | -2.762 | 1.262 | -2.467 | -1.146 |
| 14 | 1.135 | -0.79 | -2.142 | -1.146 |
| 15 | 0.258 | -1.048 | 0.743 | -1.374 |
| 16 | -0.138 | -1.353 | 0.95 | -1.253 |
| 17 | -0.878 | 1.176 | -2.189 | -1.464 |
| 18 | 0.962 | -1.129 | -0.3 | -1.146 |
| 19 | -0.322 | -0.083 | -0.15 | 0.402 |
| 20 | 0.62 | -1.312 | 0.395 | -0.945 |
| 21 | -1.762 | 1.168 | -1.048 | -1.332 |
| 22 | 0.743 | -0.107 | -1.235 | -0.345 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.81451 |
| 0.0005 | 3.94301 |
| 0.0001 | 6.31491 |