| Motif | ZBED5.H14RSNP.0.PSGIB.D |
| Gene (human) | ZBED5 (GeneCards) |
| Gene synonyms (human) | Buster1 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 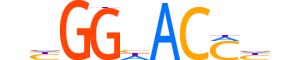 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif | ZBED5.H14RSNP.0.PSGIB.D |
| Gene (human) | ZBED5 (GeneCards) |
| Gene synonyms (human) | Buster1 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO | 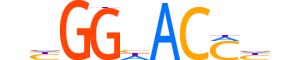 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 10 |
| Consensus | nhGGhACMhn |
| GC content | 64.21% |
| Information content (bits; total / per base) | 9.423 / 0.942 |
| Data sources | ChIP-Seq + HT-SELEX + Genomic HT-SELEX + SMiLe-Seq + PBM |
| Aligned words | 649 |
| Previous names |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|
| Overall | 2 (2) | 0.87 | 0.87 | 0.781 | 0.789 | 0.676 | 0.682 | 135.502 | 154.18 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| overall, 5 experiments | median | 0.999 | 0.999 | 0.887 | 0.898 | 0.717 | 0.75 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.999 | 0.998 | |
| Lysate, 2 experiments | median | 0.865 | 0.879 | 0.752 | 0.768 | 0.65 | 0.669 |
| best | 0.999 | 0.999 | 0.887 | 0.898 | 0.717 | 0.75 | |
| IVT, 1 experiments | median | 1.0 | 1.0 | 0.999 | 0.999 | 0.992 | 0.991 |
| best | 1.0 | 1.0 | 0.999 | 0.999 | 0.992 | 0.991 | |
| GFPIVT, 2 experiments | median | 0.833 | 0.841 | 0.792 | 0.798 | 0.774 | 0.778 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.999 | 0.998 | |
| Genomic HT-SELEX benchmarking | Centrality | pseudo-auROC | auROC | auPR | |
|---|---|---|---|---|---|
| overall, 2 experiments | median | 91.463 | 0.809 | 0.837 | 0.704 |
| best | 404.921 | 0.907 | 0.939 | 0.832 | |
| Lysate, 1 experiments | median | 74.102 | 0.727 | 0.753 | 0.675 |
| best | 108.824 | 0.741 | 0.787 | 0.71 | |
| GFPIVT, 1 experiments | median | 197.824 | 0.883 | 0.938 | 0.83 |
| best | 404.921 | 0.907 | 0.939 | 0.832 | |
| SMiLE-Seq benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.591 | 0.573 | 0.554 | 0.544 | 0.533 | 0.529 |
| best | 0.611 | 0.601 | 0.563 | 0.558 | 0.538 | 0.537 | |
| PBM benchmarking | auROC, QNZS | auPR, QNZS | auROC, SD | auPR, SD | |
|---|---|---|---|---|---|
| Overall, 3 experiments | median | 0.889 | 0.47 | 0.911 | 0.501 |
| best | 0.889 | 0.47 | 0.938 | 0.533 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | BED zinc finger {2.3.5} (TFClass) |
| TF subfamily | {2.3.5.0} (TFClass) |
| TFClass ID | TFClass: 2.3.5.0.5 |
| HGNC | HGNC:30803 |
| MGI | |
| EntrezGene (human) | GeneID:58486 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | ZBED5_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q49AG3 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | no |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 5 overall: 2 Lysate, 1 IVT, 2 GFPIVT |
| Genomic HT-SELEX | 2 overall: 1 Lysate, 0 IVT, 1 GFPIVT |
| SMiLE-Seq | 2 |
| PBM | 3 |
| PCM | ZBED5.H14RSNP.0.PSGIB.D.pcm |
| PWM | ZBED5.H14RSNP.0.PSGIB.D.pwm |
| PFM | ZBED5.H14RSNP.0.PSGIB.D.pfm |
| Threshold to P-value map | ZBED5.H14RSNP.0.PSGIB.D.thr |
| Motif in other formats | |
| JASPAR format | ZBED5.H14RSNP.0.PSGIB.D_jaspar_format.txt |
| MEME format | ZBED5.H14RSNP.0.PSGIB.D_meme_format.meme |
| Transfac format | ZBED5.H14RSNP.0.PSGIB.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 202.0 | 140.0 | 198.0 | 109.0 |
| 02 | 94.0 | 370.0 | 81.0 | 104.0 |
| 03 | 0.0 | 0.0 | 649.0 | 0.0 |
| 04 | 3.0 | 2.0 | 630.0 | 14.0 |
| 05 | 256.0 | 169.0 | 13.0 | 211.0 |
| 06 | 643.0 | 1.0 | 3.0 | 2.0 |
| 07 | 2.0 | 634.0 | 9.0 | 4.0 |
| 08 | 99.0 | 483.0 | 16.0 | 51.0 |
| 09 | 78.0 | 357.0 | 76.0 | 138.0 |
| 10 | 120.0 | 215.0 | 121.0 | 193.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.311 | 0.216 | 0.305 | 0.168 |
| 02 | 0.145 | 0.57 | 0.125 | 0.16 |
| 03 | 0.0 | 0.0 | 1.0 | 0.0 |
| 04 | 0.005 | 0.003 | 0.971 | 0.022 |
| 05 | 0.394 | 0.26 | 0.02 | 0.325 |
| 06 | 0.991 | 0.002 | 0.005 | 0.003 |
| 07 | 0.003 | 0.977 | 0.014 | 0.006 |
| 08 | 0.153 | 0.744 | 0.025 | 0.079 |
| 09 | 0.12 | 0.55 | 0.117 | 0.213 |
| 10 | 0.185 | 0.331 | 0.186 | 0.297 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.217 | -0.146 | 0.197 | -0.393 |
| 02 | -0.539 | 0.819 | -0.685 | -0.439 |
| 03 | -4.617 | -4.617 | 1.379 | -4.617 |
| 04 | -3.569 | -3.813 | 1.349 | -2.351 |
| 05 | 0.452 | 0.04 | -2.417 | 0.26 |
| 06 | 1.37 | -4.136 | -3.569 | -3.813 |
| 07 | -3.813 | 1.356 | -2.736 | -3.373 |
| 08 | -0.488 | 1.084 | -2.23 | -1.136 |
| 09 | -0.722 | 0.783 | -0.747 | -0.16 |
| 10 | -0.298 | 0.279 | -0.29 | 0.172 |
| P-value | Threshold |
|---|---|
| 0.001 | 5.21693 |
| 0.0005 | 6.03039 |
| 0.0001 | 7.3926 |