| Motif | Z780B.H14CORE.0.PSGI.A |
| Gene (human) | ZNF780B (GeneCards) |
| Gene synonyms (human) | ZNF779 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 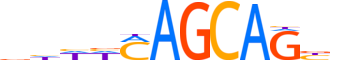 |
| Motif subtype | 0 |
| Quality | A |
| Motif | Z780B.H14CORE.0.PSGI.A |
| Gene (human) | ZNF780B (GeneCards) |
| Gene synonyms (human) | ZNF779 |
| Gene (mouse) | |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 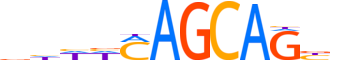 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 12 |
| Consensus | nvCTGCTKdddv |
| GC content | 50.77% |
| Information content (bits; total / per base) | 11.215 / 0.935 |
| Data sources | ChIP-Seq + HT-SELEX + Genomic HT-SELEX + SMiLe-Seq |
| Aligned words | 798 |
| Previous names |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|
| Overall | 2 (2) | 0.97 | 0.972 | 0.939 | 0.94 | 0.912 | 0.922 | 202.412 | 277.796 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| GFPIVT, 2 experiments | median | 1.0 | 1.0 | 0.986 | 0.984 | 0.892 | 0.906 |
| best | 1.0 | 1.0 | 1.0 | 0.999 | 0.996 | 0.995 | |
| Genomic HT-SELEX benchmarking | Centrality | pseudo-auROC | auROC | auPR | |
|---|---|---|---|---|---|
| GFPIVT, 2 experiments | median | 515.771 | 0.929 | 0.962 | 0.938 |
| best | 781.921 | 0.944 | 0.98 | 0.964 | |
| SMiLE-Seq benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 1 experiments | median | 0.881 | 0.862 | 0.733 | 0.729 | 0.627 | 0.641 |
| best | 0.881 | 0.862 | 0.733 | 0.729 | 0.627 | 0.641 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | ZNF283-like {2.3.3.67} (TFClass) |
| TFClass ID | TFClass: 2.3.3.67.12 |
| HGNC | HGNC:33109 |
| MGI | |
| EntrezGene (human) | GeneID:163131 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | Z780B_HUMAN |
| UniProt ID (mouse) | |
| UniProt AC (human) | Q9Y6R6 (TFClass) |
| UniProt AC (mouse) | |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 2 overall: 0 Lysate, 0 IVT, 2 GFPIVT |
| Genomic HT-SELEX | 2 overall: 0 Lysate, 0 IVT, 2 GFPIVT |
| SMiLE-Seq | 1 |
| PBM | 0 |
| PCM | Z780B.H14CORE.0.PSGI.A.pcm |
| PWM | Z780B.H14CORE.0.PSGI.A.pwm |
| PFM | Z780B.H14CORE.0.PSGI.A.pfm |
| Threshold to P-value map | Z780B.H14CORE.0.PSGI.A.thr |
| Motif in other formats | |
| JASPAR format | Z780B.H14CORE.0.PSGI.A_jaspar_format.txt |
| MEME format | Z780B.H14CORE.0.PSGI.A_meme_format.meme |
| Transfac format | Z780B.H14CORE.0.PSGI.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 212.75 | 161.75 | 188.75 | 234.75 |
| 02 | 205.75 | 134.75 | 407.75 | 49.75 |
| 03 | 40.0 | 674.0 | 29.0 | 55.0 |
| 04 | 5.0 | 2.0 | 2.0 | 789.0 |
| 05 | 1.0 | 1.0 | 796.0 | 0.0 |
| 06 | 5.0 | 793.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 798.0 |
| 08 | 52.0 | 8.0 | 562.0 | 176.0 |
| 09 | 475.0 | 89.0 | 111.0 | 123.0 |
| 10 | 470.0 | 59.0 | 128.0 | 141.0 |
| 11 | 378.0 | 89.0 | 154.0 | 177.0 |
| 12 | 205.75 | 320.75 | 150.75 | 120.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.267 | 0.203 | 0.237 | 0.294 |
| 02 | 0.258 | 0.169 | 0.511 | 0.062 |
| 03 | 0.05 | 0.845 | 0.036 | 0.069 |
| 04 | 0.006 | 0.003 | 0.003 | 0.989 |
| 05 | 0.001 | 0.001 | 0.997 | 0.0 |
| 06 | 0.006 | 0.994 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 1.0 |
| 08 | 0.065 | 0.01 | 0.704 | 0.221 |
| 09 | 0.595 | 0.112 | 0.139 | 0.154 |
| 10 | 0.589 | 0.074 | 0.16 | 0.177 |
| 11 | 0.474 | 0.112 | 0.193 | 0.222 |
| 12 | 0.258 | 0.402 | 0.189 | 0.151 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.064 | -0.208 | -0.055 | 0.161 |
| 02 | 0.031 | -0.388 | 0.711 | -1.364 |
| 03 | -1.574 | 1.212 | -1.881 | -1.267 |
| 04 | -3.406 | -4.004 | -4.004 | 1.369 |
| 05 | -4.322 | -4.322 | 1.378 | -4.791 |
| 06 | -3.406 | 1.374 | -4.791 | -4.791 |
| 07 | -4.791 | -4.791 | -4.791 | 1.38 |
| 08 | -1.321 | -3.035 | 1.03 | -0.124 |
| 09 | 0.863 | -0.797 | -0.58 | -0.478 |
| 10 | 0.852 | -1.199 | -0.439 | -0.344 |
| 11 | 0.635 | -0.797 | -0.256 | -0.119 |
| 12 | 0.031 | 0.472 | -0.278 | -0.497 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.43036 |
| 0.0005 | 5.551985 |
| 0.0001 | 7.529985 |