| Motif | Z518B.H14INVITRO.0.PSG.A |
| Gene (human) | ZNF518B (GeneCards) |
| Gene synonyms (human) | KIAA1729 |
| Gene (mouse) | Znf518b |
| Gene synonyms (mouse) | Kiaa1729, Zfp518b |
| LOGO | 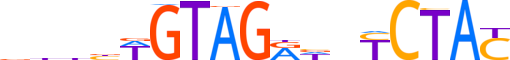 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif | Z518B.H14INVITRO.0.PSG.A |
| Gene (human) | ZNF518B (GeneCards) |
| Gene synonyms (human) | KIAA1729 |
| Gene (mouse) | Znf518b |
| Gene synonyms (mouse) | Kiaa1729, Zfp518b |
| LOGO | 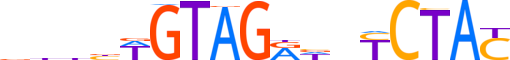 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 17 |
| Consensus | dbhbWGTAGRdnYCTAC |
| GC content | 42.85% |
| Information content (bits; total / per base) | 17.756 / 1.044 |
| Data sources | ChIP-Seq + HT-SELEX + Genomic HT-SELEX |
| Aligned words | 969 |
| Previous names |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|
| Overall | 2 (2) | 0.934 | 0.943 | 0.915 | 0.926 | 0.815 | 0.836 | 195.736 | 212.886 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| overall, 3 experiments | median | 1.0 | 0.999 | 0.999 | 0.999 | 0.992 | 0.991 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.993 | 0.993 | |
| Lysate, 2 experiments | median | 1.0 | 1.0 | 0.956 | 0.962 | 0.845 | 0.873 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.993 | 0.993 | |
| GFPIVT, 1 experiments | median | 1.0 | 0.999 | 0.999 | 0.999 | 0.992 | 0.991 |
| best | 1.0 | 0.999 | 0.999 | 0.999 | 0.992 | 0.991 | |
| Genomic HT-SELEX benchmarking | Centrality | pseudo-auROC | auROC | auPR | |
|---|---|---|---|---|---|
| overall, 3 experiments | median | 816.191 | 0.865 | 0.948 | 0.954 |
| best | 999.347 | 0.99 | 0.993 | 0.992 | |
| Lysate, 2 experiments | median | 749.515 | 0.823 | 0.931 | 0.94 |
| best | 945.337 | 0.952 | 0.993 | 0.992 | |
| GFPIVT, 1 experiments | median | 963.585 | 0.982 | 0.992 | 0.99 |
| best | 999.347 | 0.99 | 0.993 | 0.992 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | Multiple dispersed zinc fingers {2.3.4} (TFClass) |
| TF subfamily | ZNF518 {2.3.4.18} (TFClass) |
| TFClass ID | TFClass: 2.3.4.18.2 |
| HGNC | HGNC:29365 |
| MGI | MGI:2140750 |
| EntrezGene (human) | GeneID:85460 (SSTAR profile) |
| EntrezGene (mouse) | |
| UniProt ID (human) | Z518B_HUMAN |
| UniProt ID (mouse) | Z518B_MOUSE |
| UniProt AC (human) | Q9C0D4 (TFClass) |
| UniProt AC (mouse) | B2RRE4 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 3 overall: 2 Lysate, 0 IVT, 1 GFPIVT |
| Genomic HT-SELEX | 3 overall: 2 Lysate, 0 IVT, 1 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | Z518B.H14INVITRO.0.PSG.A.pcm |
| PWM | Z518B.H14INVITRO.0.PSG.A.pwm |
| PFM | Z518B.H14INVITRO.0.PSG.A.pfm |
| Threshold to P-value map | Z518B.H14INVITRO.0.PSG.A.thr |
| Motif in other formats | |
| JASPAR format | Z518B.H14INVITRO.0.PSG.A_jaspar_format.txt |
| MEME format | Z518B.H14INVITRO.0.PSG.A_meme_format.meme |
| Transfac format | Z518B.H14INVITRO.0.PSG.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 188.0 | 134.0 | 399.0 | 248.0 |
| 02 | 130.0 | 225.0 | 194.0 | 420.0 |
| 03 | 183.0 | 188.0 | 164.0 | 434.0 |
| 04 | 103.0 | 502.0 | 179.0 | 185.0 |
| 05 | 214.0 | 8.0 | 117.0 | 630.0 |
| 06 | 18.0 | 0.0 | 951.0 | 0.0 |
| 07 | 0.0 | 1.0 | 2.0 | 966.0 |
| 08 | 942.0 | 1.0 | 26.0 | 0.0 |
| 09 | 2.0 | 0.0 | 966.0 | 1.0 |
| 10 | 573.0 | 1.0 | 296.0 | 99.0 |
| 11 | 172.0 | 92.0 | 151.0 | 554.0 |
| 12 | 182.0 | 198.0 | 364.0 | 225.0 |
| 13 | 53.0 | 229.0 | 1.0 | 686.0 |
| 14 | 0.0 | 968.0 | 1.0 | 0.0 |
| 15 | 0.0 | 41.0 | 0.0 | 928.0 |
| 16 | 968.0 | 0.0 | 0.0 | 1.0 |
| 17 | 1.0 | 659.0 | 1.0 | 308.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.194 | 0.138 | 0.412 | 0.256 |
| 02 | 0.134 | 0.232 | 0.2 | 0.433 |
| 03 | 0.189 | 0.194 | 0.169 | 0.448 |
| 04 | 0.106 | 0.518 | 0.185 | 0.191 |
| 05 | 0.221 | 0.008 | 0.121 | 0.65 |
| 06 | 0.019 | 0.0 | 0.981 | 0.0 |
| 07 | 0.0 | 0.001 | 0.002 | 0.997 |
| 08 | 0.972 | 0.001 | 0.027 | 0.0 |
| 09 | 0.002 | 0.0 | 0.997 | 0.001 |
| 10 | 0.591 | 0.001 | 0.305 | 0.102 |
| 11 | 0.178 | 0.095 | 0.156 | 0.572 |
| 12 | 0.188 | 0.204 | 0.376 | 0.232 |
| 13 | 0.055 | 0.236 | 0.001 | 0.708 |
| 14 | 0.0 | 0.999 | 0.001 | 0.0 |
| 15 | 0.0 | 0.042 | 0.0 | 0.958 |
| 16 | 0.999 | 0.0 | 0.0 | 0.001 |
| 17 | 0.001 | 0.68 | 0.001 | 0.318 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.251 | -0.586 | 0.496 | 0.023 |
| 02 | -0.616 | -0.073 | -0.22 | 0.547 |
| 03 | -0.278 | -0.251 | -0.387 | 0.58 |
| 04 | -0.846 | 0.725 | -0.3 | -0.267 |
| 05 | -0.123 | -3.223 | -0.72 | 0.951 |
| 06 | -2.515 | -4.955 | 1.362 | -4.955 |
| 07 | -4.955 | -4.497 | -4.184 | 1.378 |
| 08 | 1.353 | -4.497 | -2.175 | -4.955 |
| 09 | -4.184 | -4.955 | 1.378 | -4.497 |
| 10 | 0.857 | -4.497 | 0.199 | -0.885 |
| 11 | -0.34 | -0.957 | -0.468 | 0.823 |
| 12 | -0.284 | -0.2 | 0.405 | -0.073 |
| 13 | -1.495 | -0.056 | -4.497 | 1.036 |
| 14 | -4.955 | 1.38 | -4.497 | -4.955 |
| 15 | -4.955 | -1.742 | -4.955 | 1.338 |
| 16 | 1.38 | -4.955 | -4.955 | -4.497 |
| 17 | -4.497 | 0.996 | -4.497 | 0.239 |
| P-value | Threshold |
|---|---|
| 0.001 | -0.05089 |
| 0.0005 | 1.55926 |
| 0.0001 | 5.04301 |