| Motif | THB.H14RSNP.3.S.D |
| Gene (human) | THRB (GeneCards) |
| Gene synonyms (human) | ERBA2, NR1A2, THR1 |
| Gene (mouse) | Thrb |
| Gene synonyms (mouse) | Erba2, Nr1a2 |
| LOGO |  |
| LOGO (reverse complement) | 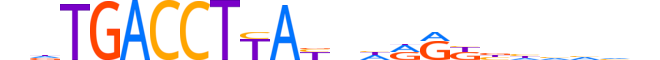 |
| Motif subtype | 3 |
| Quality | D |
| Motif | THB.H14RSNP.3.S.D |
| Gene (human) | THRB (GeneCards) |
| Gene synonyms (human) | ERBA2, NR1A2, THR1 |
| Gene (mouse) | Thrb |
| Gene synonyms (mouse) | Erba2, Nr1a2 |
| LOGO |  |
| LOGO (reverse complement) | 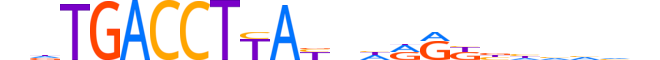 |
| Motif subtype | 3 |
| Quality | D |
| Motif length | 22 |
| Consensus | ndbddvMYYdnRTRAGGTCAhn |
| GC content | 45.4% |
| Information content (bits; total / per base) | 18.675 / 0.849 |
| Data sources | HT-SELEX |
| Aligned words | 682 |
| Previous names | THB.H12RSNP.3.S.D |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.686 | 0.704 | 0.498 | 0.512 | 0.687 | 0.693 | 1.903 | 1.961 | 63.092 | 77.244 |
| Mouse | 1 (5) | 0.558 | 0.563 | 0.414 | 0.426 | 0.547 | 0.569 | 1.231 | 1.328 | 11.886 | 17.347 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 0.943 | 0.91 | 0.911 | 0.871 | 0.845 | 0.809 |
| best | 0.998 | 0.997 | 0.994 | 0.992 | 0.974 | 0.968 | |
| Methyl HT-SELEX, 2 experiments | median | 0.972 | 0.956 | 0.954 | 0.933 | 0.912 | 0.891 |
| best | 0.998 | 0.997 | 0.994 | 0.992 | 0.974 | 0.968 | |
| Non-Methyl HT-SELEX, 4 experiments | median | 0.834 | 0.82 | 0.761 | 0.747 | 0.697 | 0.689 |
| best | 0.997 | 0.996 | 0.992 | 0.989 | 0.967 | 0.96 | |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Nuclear receptors with C4 zinc fingers {2.1} (TFClass) |
| TF family | Thyroid hormone receptor-related {2.1.2} (TFClass) |
| TF subfamily | T3R (NR1A) {2.1.2.2} (TFClass) |
| TFClass ID | TFClass: 2.1.2.2.2 |
| HGNC | HGNC:11799 |
| MGI | MGI:98743 |
| EntrezGene (human) | GeneID:7068 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21834 (SSTAR profile) |
| UniProt ID (human) | THB_HUMAN |
| UniProt ID (mouse) | THB_MOUSE |
| UniProt AC (human) | P10828 (TFClass) |
| UniProt AC (mouse) | P37242 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 1 mouse |
| HT-SELEX | 4 |
| Methyl-HT-SELEX | 2 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | THB.H14RSNP.3.S.D.pcm |
| PWM | THB.H14RSNP.3.S.D.pwm |
| PFM | THB.H14RSNP.3.S.D.pfm |
| Threshold to P-value map | THB.H14RSNP.3.S.D.thr |
| Motif in other formats | |
| JASPAR format | THB.H14RSNP.3.S.D_jaspar_format.txt |
| MEME format | THB.H14RSNP.3.S.D_meme_format.meme |
| Transfac format | THB.H14RSNP.3.S.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 205.25 | 171.25 | 122.25 | 183.25 |
| 02 | 114.5 | 90.5 | 250.5 | 226.5 |
| 03 | 91.0 | 116.0 | 155.0 | 320.0 |
| 04 | 120.0 | 81.0 | 223.0 | 258.0 |
| 05 | 225.0 | 28.0 | 196.0 | 233.0 |
| 06 | 208.0 | 184.0 | 264.0 | 26.0 |
| 07 | 227.0 | 368.0 | 50.0 | 37.0 |
| 08 | 7.0 | 453.0 | 8.0 | 214.0 |
| 09 | 51.0 | 336.0 | 18.0 | 277.0 |
| 10 | 192.0 | 61.0 | 80.0 | 349.0 |
| 11 | 218.0 | 224.0 | 113.0 | 127.0 |
| 12 | 398.0 | 71.0 | 166.0 | 47.0 |
| 13 | 4.0 | 7.0 | 1.0 | 670.0 |
| 14 | 489.0 | 11.0 | 171.0 | 11.0 |
| 15 | 682.0 | 0.0 | 0.0 | 0.0 |
| 16 | 0.0 | 1.0 | 681.0 | 0.0 |
| 17 | 0.0 | 0.0 | 682.0 | 0.0 |
| 18 | 0.0 | 0.0 | 0.0 | 682.0 |
| 19 | 1.0 | 680.0 | 1.0 | 0.0 |
| 20 | 672.0 | 1.0 | 9.0 | 0.0 |
| 21 | 114.5 | 267.5 | 57.5 | 242.5 |
| 22 | 98.5 | 181.5 | 231.5 | 170.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.301 | 0.251 | 0.179 | 0.269 |
| 02 | 0.168 | 0.133 | 0.367 | 0.332 |
| 03 | 0.133 | 0.17 | 0.227 | 0.469 |
| 04 | 0.176 | 0.119 | 0.327 | 0.378 |
| 05 | 0.33 | 0.041 | 0.287 | 0.342 |
| 06 | 0.305 | 0.27 | 0.387 | 0.038 |
| 07 | 0.333 | 0.54 | 0.073 | 0.054 |
| 08 | 0.01 | 0.664 | 0.012 | 0.314 |
| 09 | 0.075 | 0.493 | 0.026 | 0.406 |
| 10 | 0.282 | 0.089 | 0.117 | 0.512 |
| 11 | 0.32 | 0.328 | 0.166 | 0.186 |
| 12 | 0.584 | 0.104 | 0.243 | 0.069 |
| 13 | 0.006 | 0.01 | 0.001 | 0.982 |
| 14 | 0.717 | 0.016 | 0.251 | 0.016 |
| 15 | 1.0 | 0.0 | 0.0 | 0.0 |
| 16 | 0.0 | 0.001 | 0.999 | 0.0 |
| 17 | 0.0 | 0.0 | 1.0 | 0.0 |
| 18 | 0.0 | 0.0 | 0.0 | 1.0 |
| 19 | 0.001 | 0.997 | 0.001 | 0.0 |
| 20 | 0.985 | 0.001 | 0.013 | 0.0 |
| 21 | 0.168 | 0.392 | 0.084 | 0.356 |
| 22 | 0.144 | 0.266 | 0.339 | 0.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.184 | 0.004 | -0.329 | 0.071 |
| 02 | -0.394 | -0.625 | 0.382 | 0.282 |
| 03 | -0.62 | -0.381 | -0.094 | 0.625 |
| 04 | -0.347 | -0.734 | 0.266 | 0.411 |
| 05 | 0.275 | -1.759 | 0.138 | 0.31 |
| 06 | 0.197 | 0.076 | 0.434 | -1.829 |
| 07 | 0.284 | 0.764 | -1.204 | -1.494 |
| 08 | -2.993 | 0.971 | -2.883 | 0.225 |
| 09 | -1.185 | 0.674 | -2.171 | 0.482 |
| 10 | 0.118 | -1.011 | -0.746 | 0.711 |
| 11 | 0.244 | 0.271 | -0.407 | -0.291 |
| 12 | 0.842 | -0.863 | -0.026 | -1.264 |
| 13 | -3.42 | -2.993 | -4.181 | 1.361 |
| 14 | 1.047 | -2.612 | 0.003 | -2.612 |
| 15 | 1.379 | -4.659 | -4.659 | -4.659 |
| 16 | -4.659 | -4.181 | 1.378 | -4.659 |
| 17 | -4.659 | -4.659 | 1.379 | -4.659 |
| 18 | -4.659 | -4.659 | -4.659 | 1.379 |
| 19 | -4.181 | 1.376 | -4.181 | -4.659 |
| 20 | 1.364 | -4.181 | -2.784 | -4.659 |
| 21 | -0.394 | 0.447 | -1.068 | 0.349 |
| 22 | -0.542 | 0.062 | 0.303 | 0.0 |
| P-value | Threshold |
|---|---|
| 0.001 | -0.19694 |
| 0.0005 | 1.38221 |
| 0.0001 | 4.77196 |