| Motif | TGIF1.H14CORE.1.S.B |
| Gene (human) | TGIF1 (GeneCards) |
| Gene synonyms (human) | TGIF |
| Gene (mouse) | Tgif1 |
| Gene synonyms (mouse) | Tgif |
| LOGO | 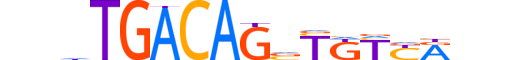 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif | TGIF1.H14CORE.1.S.B |
| Gene (human) | TGIF1 (GeneCards) |
| Gene synonyms (human) | TGIF |
| Gene (mouse) | Tgif1 |
| Gene synonyms (mouse) | Tgif |
| LOGO | 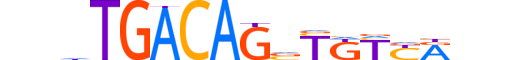 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 17 |
| Consensus | nnhTGACAGvTKYYRnn |
| GC content | 45.36% |
| Information content (bits; total / per base) | 15.572 / 0.916 |
| Data sources | HT-SELEX |
| Aligned words | 511 |
| Previous names | TGIF1.H12CORE.1.S.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 3 (17) | 0.81 | 0.877 | 0.655 | 0.743 | 0.767 | 0.853 | 2.258 | 3.006 | 111.569 | 306.678 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.723 | 0.696 | 0.678 | 0.651 | 0.623 | 0.609 |
| best | 0.962 | 0.942 | 0.941 | 0.912 | 0.881 | 0.855 | |
| Methyl HT-SELEX, 1 experiments | median | 0.868 | 0.821 | 0.815 | 0.763 | 0.722 | 0.695 |
| best | 0.868 | 0.821 | 0.815 | 0.763 | 0.722 | 0.695 | |
| Non-Methyl HT-SELEX, 3 experiments | median | 0.579 | 0.571 | 0.541 | 0.539 | 0.523 | 0.524 |
| best | 0.962 | 0.942 | 0.941 | 0.912 | 0.881 | 0.855 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | TALE-type HD {3.1.4} (TFClass) |
| TF subfamily | TGIF {3.1.4.6} (TFClass) |
| TFClass ID | TFClass: 3.1.4.6.1 |
| HGNC | HGNC:11776 |
| MGI | MGI:1194497 |
| EntrezGene (human) | GeneID:7050 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21815 (SSTAR profile) |
| UniProt ID (human) | TGIF1_HUMAN |
| UniProt ID (mouse) | TGIF1_MOUSE |
| UniProt AC (human) | Q15583 (TFClass) |
| UniProt AC (mouse) | P70284 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 3 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 1 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | TGIF1.H14CORE.1.S.B.pcm |
| PWM | TGIF1.H14CORE.1.S.B.pwm |
| PFM | TGIF1.H14CORE.1.S.B.pfm |
| Threshold to P-value map | TGIF1.H14CORE.1.S.B.thr |
| Motif in other formats | |
| JASPAR format | TGIF1.H14CORE.1.S.B_jaspar_format.txt |
| MEME format | TGIF1.H14CORE.1.S.B_meme_format.meme |
| Transfac format | TGIF1.H14CORE.1.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 156.25 | 97.25 | 121.25 | 136.25 |
| 02 | 155.0 | 73.0 | 143.0 | 140.0 |
| 03 | 166.0 | 110.0 | 57.0 | 178.0 |
| 04 | 0.0 | 0.0 | 0.0 | 511.0 |
| 05 | 0.0 | 0.0 | 511.0 | 0.0 |
| 06 | 494.0 | 0.0 | 0.0 | 17.0 |
| 07 | 0.0 | 511.0 | 0.0 | 0.0 |
| 08 | 511.0 | 0.0 | 0.0 | 0.0 |
| 09 | 6.0 | 11.0 | 440.0 | 54.0 |
| 10 | 78.0 | 227.0 | 169.0 | 37.0 |
| 11 | 19.0 | 56.0 | 26.0 | 410.0 |
| 12 | 50.0 | 27.0 | 380.0 | 54.0 |
| 13 | 43.0 | 44.0 | 30.0 | 394.0 |
| 14 | 41.0 | 347.0 | 51.0 | 72.0 |
| 15 | 360.0 | 26.0 | 63.0 | 62.0 |
| 16 | 152.0 | 151.0 | 56.0 | 152.0 |
| 17 | 171.75 | 110.75 | 101.75 | 126.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.306 | 0.19 | 0.237 | 0.267 |
| 02 | 0.303 | 0.143 | 0.28 | 0.274 |
| 03 | 0.325 | 0.215 | 0.112 | 0.348 |
| 04 | 0.0 | 0.0 | 0.0 | 1.0 |
| 05 | 0.0 | 0.0 | 1.0 | 0.0 |
| 06 | 0.967 | 0.0 | 0.0 | 0.033 |
| 07 | 0.0 | 1.0 | 0.0 | 0.0 |
| 08 | 1.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.012 | 0.022 | 0.861 | 0.106 |
| 10 | 0.153 | 0.444 | 0.331 | 0.072 |
| 11 | 0.037 | 0.11 | 0.051 | 0.802 |
| 12 | 0.098 | 0.053 | 0.744 | 0.106 |
| 13 | 0.084 | 0.086 | 0.059 | 0.771 |
| 14 | 0.08 | 0.679 | 0.1 | 0.141 |
| 15 | 0.705 | 0.051 | 0.123 | 0.121 |
| 16 | 0.297 | 0.295 | 0.11 | 0.297 |
| 17 | 0.336 | 0.217 | 0.199 | 0.248 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.199 | -0.269 | -0.052 | 0.064 |
| 02 | 0.191 | -0.551 | 0.111 | 0.091 |
| 03 | 0.259 | -0.148 | -0.792 | 0.328 |
| 04 | -4.418 | -4.418 | -4.418 | 1.377 |
| 05 | -4.418 | -4.418 | 1.377 | -4.418 |
| 06 | 1.343 | -4.418 | -4.418 | -1.941 |
| 07 | -4.418 | 1.377 | -4.418 | -4.418 |
| 08 | 1.377 | -4.418 | -4.418 | -4.418 |
| 09 | -2.839 | -2.332 | 1.228 | -0.845 |
| 10 | -0.486 | 0.57 | 0.277 | -1.21 |
| 11 | -1.839 | -0.809 | -1.546 | 1.158 |
| 12 | -0.919 | -1.51 | 1.082 | -0.845 |
| 13 | -1.065 | -1.043 | -1.41 | 1.118 |
| 14 | -1.111 | 0.992 | -0.9 | -0.564 |
| 15 | 1.028 | -1.546 | -0.695 | -0.71 |
| 16 | 0.172 | 0.165 | -0.809 | 0.172 |
| 17 | 0.293 | -0.141 | -0.224 | -0.008 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.46041 |
| 0.0005 | 3.78526 |
| 0.0001 | 6.51576 |