| Motif | TBX5.H14RSNP.2.S.D |
| Gene (human) | TBX5 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Tbx5 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 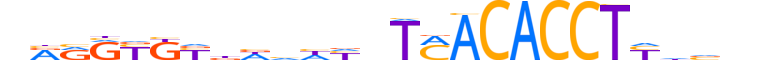 |
| Motif subtype | 2 |
| Quality | D |
| Motif | TBX5.H14RSNP.2.S.D |
| Gene (human) | TBX5 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Tbx5 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 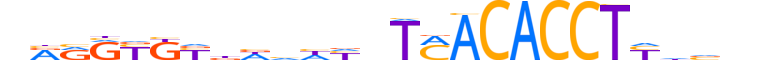 |
| Motif subtype | 2 |
| Quality | D |
| Motif length | 26 |
| Consensus | nnbdRAGGTGTKAndddhvRMRYYYn |
| GC content | 45.66% |
| Information content (bits; total / per base) | 19.813 / 0.762 |
| Data sources | HT-SELEX |
| Aligned words | 269 |
| Previous names | TBX5.H12RSNP.2.S.D |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 8 (52) | 0.609 | 0.641 | 0.415 | 0.439 | 0.574 | 0.599 | 1.25 | 1.36 | 13.0 | 26.409 |
| Mouse | 9 (57) | 0.728 | 0.842 | 0.578 | 0.75 | 0.68 | 0.8 | 1.904 | 3.004 | 43.658 | 109.119 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.743 | 0.733 | 0.645 | 0.64 | 0.587 | 0.589 |
| best | 0.811 | 0.805 | 0.688 | 0.685 | 0.613 | 0.618 | |
| TF superclass | Immunoglobulin fold {6} (TFClass) |
| TF class | T-Box factors {6.5} (TFClass) |
| TF family | TBX2-related {6.5.4} (TFClass) |
| TF subfamily | {6.5.4.0} (TFClass) |
| TFClass ID | TFClass: 6.5.4.0.4 |
| HGNC | HGNC:11604 |
| MGI | MGI:102541 |
| EntrezGene (human) | GeneID:6910 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21388 (SSTAR profile) |
| UniProt ID (human) | TBX5_HUMAN |
| UniProt ID (mouse) | TBX5_MOUSE |
| UniProt AC (human) | Q99593 (TFClass) |
| UniProt AC (mouse) | P70326 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 8 human, 9 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | TBX5.H14RSNP.2.S.D.pcm |
| PWM | TBX5.H14RSNP.2.S.D.pwm |
| PFM | TBX5.H14RSNP.2.S.D.pfm |
| Threshold to P-value map | TBX5.H14RSNP.2.S.D.thr |
| Motif in other formats | |
| JASPAR format | TBX5.H14RSNP.2.S.D_jaspar_format.txt |
| MEME format | TBX5.H14RSNP.2.S.D_meme_format.meme |
| Transfac format | TBX5.H14RSNP.2.S.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 45.25 | 64.25 | 98.25 | 61.25 |
| 02 | 76.5 | 48.5 | 90.5 | 53.5 |
| 03 | 22.0 | 66.0 | 118.0 | 63.0 |
| 04 | 93.0 | 27.0 | 66.0 | 83.0 |
| 05 | 162.0 | 4.0 | 61.0 | 42.0 |
| 06 | 264.0 | 0.0 | 5.0 | 0.0 |
| 07 | 0.0 | 1.0 | 268.0 | 0.0 |
| 08 | 0.0 | 0.0 | 269.0 | 0.0 |
| 09 | 0.0 | 0.0 | 1.0 | 268.0 |
| 10 | 0.0 | 0.0 | 269.0 | 0.0 |
| 11 | 5.0 | 2.0 | 9.0 | 253.0 |
| 12 | 1.0 | 11.0 | 164.0 | 93.0 |
| 13 | 244.0 | 6.0 | 8.0 | 11.0 |
| 14 | 76.0 | 52.0 | 57.0 | 84.0 |
| 15 | 157.0 | 24.0 | 47.0 | 41.0 |
| 16 | 92.0 | 10.0 | 46.0 | 121.0 |
| 17 | 62.0 | 28.0 | 75.0 | 104.0 |
| 18 | 46.0 | 37.0 | 30.0 | 156.0 |
| 19 | 97.0 | 98.0 | 49.0 | 25.0 |
| 20 | 175.0 | 30.0 | 34.0 | 30.0 |
| 21 | 36.0 | 199.0 | 23.0 | 11.0 |
| 22 | 183.0 | 33.0 | 37.0 | 16.0 |
| 23 | 21.0 | 201.0 | 23.0 | 24.0 |
| 24 | 7.0 | 165.0 | 42.0 | 55.0 |
| 25 | 24.5 | 40.5 | 30.5 | 173.5 |
| 26 | 59.0 | 80.0 | 46.0 | 84.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.168 | 0.239 | 0.365 | 0.228 |
| 02 | 0.284 | 0.18 | 0.336 | 0.199 |
| 03 | 0.082 | 0.245 | 0.439 | 0.234 |
| 04 | 0.346 | 0.1 | 0.245 | 0.309 |
| 05 | 0.602 | 0.015 | 0.227 | 0.156 |
| 06 | 0.981 | 0.0 | 0.019 | 0.0 |
| 07 | 0.0 | 0.004 | 0.996 | 0.0 |
| 08 | 0.0 | 0.0 | 1.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.004 | 0.996 |
| 10 | 0.0 | 0.0 | 1.0 | 0.0 |
| 11 | 0.019 | 0.007 | 0.033 | 0.941 |
| 12 | 0.004 | 0.041 | 0.61 | 0.346 |
| 13 | 0.907 | 0.022 | 0.03 | 0.041 |
| 14 | 0.283 | 0.193 | 0.212 | 0.312 |
| 15 | 0.584 | 0.089 | 0.175 | 0.152 |
| 16 | 0.342 | 0.037 | 0.171 | 0.45 |
| 17 | 0.23 | 0.104 | 0.279 | 0.387 |
| 18 | 0.171 | 0.138 | 0.112 | 0.58 |
| 19 | 0.361 | 0.364 | 0.182 | 0.093 |
| 20 | 0.651 | 0.112 | 0.126 | 0.112 |
| 21 | 0.134 | 0.74 | 0.086 | 0.041 |
| 22 | 0.68 | 0.123 | 0.138 | 0.059 |
| 23 | 0.078 | 0.747 | 0.086 | 0.089 |
| 24 | 0.026 | 0.613 | 0.156 | 0.204 |
| 25 | 0.091 | 0.151 | 0.113 | 0.645 |
| 26 | 0.219 | 0.297 | 0.171 | 0.312 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.386 | -0.045 | 0.373 | -0.091 |
| 02 | 0.126 | -0.319 | 0.292 | -0.224 |
| 03 | -1.076 | -0.018 | 0.553 | -0.064 |
| 04 | 0.319 | -0.883 | -0.018 | 0.207 |
| 05 | 0.867 | -2.543 | -0.095 | -0.459 |
| 06 | 1.352 | -3.893 | -2.373 | -3.893 |
| 07 | -3.893 | -3.354 | 1.367 | -3.893 |
| 08 | -3.893 | -3.893 | 1.371 | -3.893 |
| 09 | -3.893 | -3.893 | -3.354 | 1.367 |
| 10 | -3.893 | -3.893 | 1.371 | -3.893 |
| 11 | -2.373 | -3.006 | -1.887 | 1.31 |
| 12 | -3.354 | -1.711 | 0.879 | 0.319 |
| 13 | 1.274 | -2.228 | -1.988 | -1.711 |
| 14 | 0.12 | -0.251 | -0.162 | 0.218 |
| 15 | 0.836 | -0.994 | -0.35 | -0.482 |
| 16 | 0.308 | -1.796 | -0.37 | 0.578 |
| 17 | -0.08 | -0.848 | 0.107 | 0.429 |
| 18 | -0.37 | -0.581 | -0.782 | 0.83 |
| 19 | 0.36 | 0.37 | -0.309 | -0.956 |
| 20 | 0.944 | -0.782 | -0.662 | -0.782 |
| 21 | -0.607 | 1.071 | -1.034 | -1.711 |
| 22 | 0.988 | -0.691 | -0.581 | -1.373 |
| 23 | -1.12 | 1.081 | -1.034 | -0.994 |
| 24 | -2.101 | 0.885 | -0.459 | -0.197 |
| 25 | -0.975 | -0.494 | -0.766 | 0.935 |
| 26 | -0.128 | 0.17 | -0.37 | 0.218 |
| P-value | Threshold |
|---|---|
| 0.001 | 0.99816 |
| 0.0005 | 2.36791 |
| 0.0001 | 5.25871 |