| Motif | TBX19.H14RSNP.0.PS.D |
| Gene (human) | TBX19 (GeneCards) |
| Gene synonyms (human) | TPIT |
| Gene (mouse) | Tbx19 |
| Gene synonyms (mouse) | Tpit |
| LOGO |  |
| LOGO (reverse complement) | 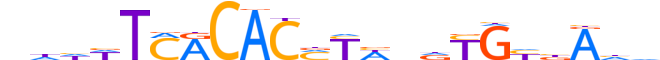 |
| Motif subtype | 0 |
| Quality | D |
| Motif | TBX19.H14RSNP.0.PS.D |
| Gene (human) | TBX19 (GeneCards) |
| Gene synonyms (human) | TPIT |
| Gene (mouse) | Tbx19 |
| Gene synonyms (mouse) | Tpit |
| LOGO |  |
| LOGO (reverse complement) | 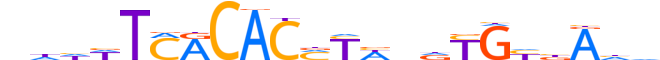 |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 22 |
| Consensus | ddTvdCRhnWRKGTGYKAWddn |
| GC content | 42.51% |
| Information content (bits; total / per base) | 16.294 / 0.741 |
| Data sources | ChIP-Seq + HT-SELEX |
| Aligned words | 7617 |
| Previous names | TBX19.H12RSNP.0.PS.D |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 2 (9) | 0.86 | 0.87 | 0.786 | 0.79 | 0.821 | 0.83 | 3.05 | 3.091 | 284.699 | 325.284 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.998 | 0.997 | 0.991 | 0.988 | 0.95 | 0.949 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.974 | 0.966 | |
| Methyl HT-SELEX, 1 experiments | median | 0.998 | 0.996 | 0.991 | 0.988 | 0.97 | 0.962 |
| best | 0.998 | 0.996 | 0.991 | 0.988 | 0.97 | 0.962 | |
| Non-Methyl HT-SELEX, 3 experiments | median | 0.999 | 0.998 | 0.992 | 0.988 | 0.929 | 0.936 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.974 | 0.966 | |
| TF superclass | Immunoglobulin fold {6} (TFClass) |
| TF class | T-Box factors {6.5} (TFClass) |
| TF family | Brachyury-related {6.5.1} (TFClass) |
| TF subfamily | {6.5.1.0} (TFClass) |
| TFClass ID | TFClass: 6.5.1.0.2 |
| HGNC | HGNC:11596 |
| MGI | MGI:1891158 |
| EntrezGene (human) | GeneID:9095 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:83993 (SSTAR profile) |
| UniProt ID (human) | TBX19_HUMAN |
| UniProt ID (mouse) | TBX19_MOUSE |
| UniProt AC (human) | O60806 (TFClass) |
| UniProt AC (mouse) | Q99ME7 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 2 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 1 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | TBX19.H14RSNP.0.PS.D.pcm |
| PWM | TBX19.H14RSNP.0.PS.D.pwm |
| PFM | TBX19.H14RSNP.0.PS.D.pfm |
| Threshold to P-value map | TBX19.H14RSNP.0.PS.D.thr |
| Motif in other formats | |
| JASPAR format | TBX19.H14RSNP.0.PS.D_jaspar_format.txt |
| MEME format | TBX19.H14RSNP.0.PS.D_meme_format.meme |
| Transfac format | TBX19.H14RSNP.0.PS.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1775.5 | 1269.5 | 1525.5 | 3046.5 |
| 02 | 1306.0 | 1024.0 | 1437.0 | 3850.0 |
| 03 | 305.0 | 480.0 | 265.0 | 6567.0 |
| 04 | 2129.0 | 3535.0 | 1370.0 | 583.0 |
| 05 | 3839.0 | 390.0 | 2502.0 | 886.0 |
| 06 | 295.0 | 6701.0 | 269.0 | 352.0 |
| 07 | 5727.0 | 286.0 | 1349.0 | 255.0 |
| 08 | 919.0 | 4067.0 | 772.0 | 1859.0 |
| 09 | 1969.0 | 2151.0 | 2274.0 | 1223.0 |
| 10 | 1318.0 | 721.0 | 919.0 | 4659.0 |
| 11 | 5886.0 | 269.0 | 948.0 | 514.0 |
| 12 | 813.0 | 947.0 | 4731.0 | 1126.0 |
| 13 | 563.0 | 68.0 | 6857.0 | 129.0 |
| 14 | 3.0 | 198.0 | 83.0 | 7333.0 |
| 15 | 7.0 | 1.0 | 7603.0 | 6.0 |
| 16 | 279.0 | 1627.0 | 97.0 | 5614.0 |
| 17 | 119.0 | 307.0 | 5798.0 | 1393.0 |
| 18 | 7363.0 | 67.0 | 132.0 | 55.0 |
| 19 | 4692.0 | 844.0 | 838.0 | 1243.0 |
| 20 | 3934.0 | 932.0 | 962.0 | 1789.0 |
| 21 | 2571.75 | 735.75 | 909.75 | 3399.75 |
| 22 | 2014.75 | 1562.75 | 1414.75 | 2624.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.233 | 0.167 | 0.2 | 0.4 |
| 02 | 0.171 | 0.134 | 0.189 | 0.505 |
| 03 | 0.04 | 0.063 | 0.035 | 0.862 |
| 04 | 0.28 | 0.464 | 0.18 | 0.077 |
| 05 | 0.504 | 0.051 | 0.328 | 0.116 |
| 06 | 0.039 | 0.88 | 0.035 | 0.046 |
| 07 | 0.752 | 0.038 | 0.177 | 0.033 |
| 08 | 0.121 | 0.534 | 0.101 | 0.244 |
| 09 | 0.259 | 0.282 | 0.299 | 0.161 |
| 10 | 0.173 | 0.095 | 0.121 | 0.612 |
| 11 | 0.773 | 0.035 | 0.124 | 0.067 |
| 12 | 0.107 | 0.124 | 0.621 | 0.148 |
| 13 | 0.074 | 0.009 | 0.9 | 0.017 |
| 14 | 0.0 | 0.026 | 0.011 | 0.963 |
| 15 | 0.001 | 0.0 | 0.998 | 0.001 |
| 16 | 0.037 | 0.214 | 0.013 | 0.737 |
| 17 | 0.016 | 0.04 | 0.761 | 0.183 |
| 18 | 0.967 | 0.009 | 0.017 | 0.007 |
| 19 | 0.616 | 0.111 | 0.11 | 0.163 |
| 20 | 0.516 | 0.122 | 0.126 | 0.235 |
| 21 | 0.338 | 0.097 | 0.119 | 0.446 |
| 22 | 0.265 | 0.205 | 0.186 | 0.345 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.07 | -0.405 | -0.221 | 0.469 |
| 02 | -0.377 | -0.619 | -0.281 | 0.703 |
| 03 | -1.825 | -1.375 | -1.965 | 1.237 |
| 04 | 0.111 | 0.618 | -0.329 | -1.181 |
| 05 | 0.701 | -1.581 | 0.273 | -0.764 |
| 06 | -1.858 | 1.257 | -1.95 | -1.683 |
| 07 | 1.1 | -1.889 | -0.344 | -2.003 |
| 08 | -0.727 | 0.758 | -0.901 | -0.024 |
| 09 | 0.033 | 0.122 | 0.177 | -0.442 |
| 10 | -0.367 | -0.969 | -0.727 | 0.894 |
| 11 | 1.128 | -1.95 | -0.696 | -1.306 |
| 12 | -0.85 | -0.697 | 0.909 | -0.525 |
| 13 | -1.216 | -3.301 | 1.28 | -2.676 |
| 14 | -5.898 | -2.254 | -3.108 | 1.347 |
| 15 | -5.33 | -6.379 | 1.384 | -5.445 |
| 16 | -1.914 | -0.157 | -2.956 | 1.08 |
| 17 | -2.755 | -1.819 | 1.113 | -0.312 |
| 18 | 1.352 | -3.316 | -2.653 | -3.506 |
| 19 | 0.901 | -0.812 | -0.819 | -0.426 |
| 20 | 0.725 | -0.713 | -0.682 | -0.062 |
| 21 | 0.3 | -0.949 | -0.737 | 0.579 |
| 22 | 0.056 | -0.197 | -0.297 | 0.321 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.51281 |
| 0.0005 | 3.71926 |
| 0.0001 | 6.23126 |