| Motif | SIX1.H14RSNP.0.P.B |
| Gene (human) | SIX1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six1 |
| Gene synonyms (mouse) | |
| LOGO | 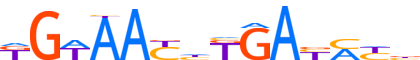 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | SIX1.H14RSNP.0.P.B |
| Gene (human) | SIX1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six1 |
| Gene synonyms (mouse) | |
| LOGO | 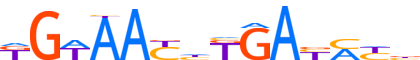 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 14 |
| Consensus | KGWAAYhKGAYMYb |
| GC content | 39.95% |
| Information content (bits; total / per base) | 12.668 / 0.905 |
| Data sources | ChIP-Seq |
| Aligned words | 145 |
| Previous names | SIX1.H12RSNP.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 3 (18) | 0.928 | 0.942 | 0.873 | 0.901 | 0.924 | 0.943 | 4.101 | 4.672 | 262.77 | 331.553 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.72 | 0.691 | 0.651 | 0.63 | 0.59 | 0.587 |
| best | 0.87 | 0.847 | 0.733 | 0.723 | 0.645 | 0.642 | |
| Methyl HT-SELEX, 2 experiments | median | 0.72 | 0.696 | 0.64 | 0.627 | 0.58 | 0.581 |
| best | 0.87 | 0.847 | 0.732 | 0.723 | 0.632 | 0.64 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.72 | 0.691 | 0.651 | 0.63 | 0.597 | 0.588 |
| best | 0.85 | 0.825 | 0.733 | 0.715 | 0.645 | 0.642 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.729 | 0.47 | 0.788 | 0.625 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | HD-SINE {3.1.6} (TFClass) |
| TF subfamily | SIX1-like {3.1.6.1} (TFClass) |
| TFClass ID | TFClass: 3.1.6.1.1 |
| HGNC | HGNC:10887 |
| MGI | MGI:102780 |
| EntrezGene (human) | GeneID:6495 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20471 (SSTAR profile) |
| UniProt ID (human) | SIX1_HUMAN |
| UniProt ID (mouse) | SIX1_MOUSE |
| UniProt AC (human) | Q15475 (TFClass) |
| UniProt AC (mouse) | Q62231 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 3 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | SIX1.H14RSNP.0.P.B.pcm |
| PWM | SIX1.H14RSNP.0.P.B.pwm |
| PFM | SIX1.H14RSNP.0.P.B.pfm |
| Threshold to P-value map | SIX1.H14RSNP.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | SIX1.H14RSNP.0.P.B_jaspar_format.txt |
| MEME format | SIX1.H14RSNP.0.P.B_meme_format.meme |
| Transfac format | SIX1.H14RSNP.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 26.0 | 2.0 | 30.0 | 87.0 |
| 02 | 2.0 | 2.0 | 139.0 | 2.0 |
| 03 | 53.0 | 5.0 | 4.0 | 83.0 |
| 04 | 133.0 | 4.0 | 1.0 | 7.0 |
| 05 | 140.0 | 1.0 | 2.0 | 2.0 |
| 06 | 4.0 | 89.0 | 8.0 | 44.0 |
| 07 | 32.0 | 59.0 | 10.0 | 44.0 |
| 08 | 8.0 | 11.0 | 20.0 | 106.0 |
| 09 | 18.0 | 2.0 | 123.0 | 2.0 |
| 10 | 142.0 | 0.0 | 1.0 | 2.0 |
| 11 | 6.0 | 29.0 | 27.0 | 83.0 |
| 12 | 72.0 | 65.0 | 6.0 | 2.0 |
| 13 | 12.0 | 69.0 | 6.0 | 58.0 |
| 14 | 13.0 | 62.0 | 34.0 | 36.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.179 | 0.014 | 0.207 | 0.6 |
| 02 | 0.014 | 0.014 | 0.959 | 0.014 |
| 03 | 0.366 | 0.034 | 0.028 | 0.572 |
| 04 | 0.917 | 0.028 | 0.007 | 0.048 |
| 05 | 0.966 | 0.007 | 0.014 | 0.014 |
| 06 | 0.028 | 0.614 | 0.055 | 0.303 |
| 07 | 0.221 | 0.407 | 0.069 | 0.303 |
| 08 | 0.055 | 0.076 | 0.138 | 0.731 |
| 09 | 0.124 | 0.014 | 0.848 | 0.014 |
| 10 | 0.979 | 0.0 | 0.007 | 0.014 |
| 11 | 0.041 | 0.2 | 0.186 | 0.572 |
| 12 | 0.497 | 0.448 | 0.041 | 0.014 |
| 13 | 0.083 | 0.476 | 0.041 | 0.4 |
| 14 | 0.09 | 0.428 | 0.234 | 0.248 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.319 | -2.447 | -0.182 | 0.856 |
| 02 | -2.447 | -2.447 | 1.319 | -2.447 |
| 03 | 0.369 | -1.793 | -1.967 | 0.81 |
| 04 | 1.275 | -1.967 | -2.816 | -1.515 |
| 05 | 1.326 | -2.816 | -2.447 | -2.447 |
| 06 | -1.967 | 0.878 | -1.4 | 0.188 |
| 07 | -0.12 | 0.474 | -1.204 | 0.188 |
| 08 | -1.4 | -1.119 | -0.568 | 1.051 |
| 09 | -0.667 | -2.447 | 1.198 | -2.447 |
| 10 | 1.34 | -3.406 | -2.816 | -2.447 |
| 11 | -1.644 | -0.215 | -0.283 | 0.81 |
| 12 | 0.67 | 0.569 | -1.644 | -2.447 |
| 13 | -1.041 | 0.628 | -1.644 | 0.457 |
| 14 | -0.968 | 0.523 | -0.062 | -0.007 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.23191 |
| 0.0005 | 5.17826 |
| 0.0001 | 7.14796 |