| Motif | SIX1.H14INVIVO.0.P.B |
| Gene (human) | SIX1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six1 |
| Gene synonyms (mouse) | |
| LOGO | 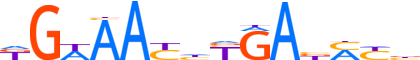 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | SIX1.H14INVIVO.0.P.B |
| Gene (human) | SIX1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Six1 |
| Gene synonyms (mouse) | |
| LOGO | 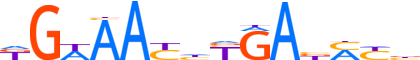 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 14 |
| Consensus | KGWAAYhYGAbMYb |
| GC content | 40.45% |
| Information content (bits; total / per base) | 12.508 / 0.893 |
| Data sources | ChIP-Seq |
| Aligned words | 1007 |
| Previous names | SIX1.H12INVIVO.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 3 (18) | 0.929 | 0.955 | 0.882 | 0.919 | 0.93 | 0.956 | 4.118 | 4.527 | 272.565 | 335.745 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.692 | 0.671 | 0.624 | 0.611 | 0.572 | 0.572 |
| best | 0.867 | 0.843 | 0.721 | 0.715 | 0.624 | 0.633 | |
| Methyl HT-SELEX, 2 experiments | median | 0.699 | 0.681 | 0.619 | 0.614 | 0.564 | 0.57 |
| best | 0.867 | 0.843 | 0.721 | 0.715 | 0.619 | 0.633 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.692 | 0.671 | 0.624 | 0.611 | 0.574 | 0.572 |
| best | 0.837 | 0.813 | 0.712 | 0.7 | 0.624 | 0.628 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.747 | 0.434 | 0.76 | 0.616 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | HD-SINE {3.1.6} (TFClass) |
| TF subfamily | SIX1-like {3.1.6.1} (TFClass) |
| TFClass ID | TFClass: 3.1.6.1.1 |
| HGNC | HGNC:10887 |
| MGI | MGI:102780 |
| EntrezGene (human) | GeneID:6495 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:20471 (SSTAR profile) |
| UniProt ID (human) | SIX1_HUMAN |
| UniProt ID (mouse) | SIX1_MOUSE |
| UniProt AC (human) | Q15475 (TFClass) |
| UniProt AC (mouse) | Q62231 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 3 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | SIX1.H14INVIVO.0.P.B.pcm |
| PWM | SIX1.H14INVIVO.0.P.B.pwm |
| PFM | SIX1.H14INVIVO.0.P.B.pfm |
| Threshold to P-value map | SIX1.H14INVIVO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | SIX1.H14INVIVO.0.P.B_jaspar_format.txt |
| MEME format | SIX1.H14INVIVO.0.P.B_meme_format.meme |
| Transfac format | SIX1.H14INVIVO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 207.0 | 20.0 | 209.0 | 571.0 |
| 02 | 6.0 | 3.0 | 991.0 | 7.0 |
| 03 | 508.0 | 43.0 | 43.0 | 413.0 |
| 04 | 919.0 | 50.0 | 8.0 | 30.0 |
| 05 | 1003.0 | 2.0 | 2.0 | 0.0 |
| 06 | 34.0 | 544.0 | 49.0 | 380.0 |
| 07 | 192.0 | 430.0 | 65.0 | 320.0 |
| 08 | 94.0 | 106.0 | 105.0 | 702.0 |
| 09 | 116.0 | 10.0 | 837.0 | 44.0 |
| 10 | 1000.0 | 0.0 | 6.0 | 1.0 |
| 11 | 64.0 | 183.0 | 278.0 | 482.0 |
| 12 | 532.0 | 402.0 | 20.0 | 53.0 |
| 13 | 83.0 | 590.0 | 56.0 | 278.0 |
| 14 | 98.0 | 412.0 | 238.0 | 259.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.206 | 0.02 | 0.208 | 0.567 |
| 02 | 0.006 | 0.003 | 0.984 | 0.007 |
| 03 | 0.504 | 0.043 | 0.043 | 0.41 |
| 04 | 0.913 | 0.05 | 0.008 | 0.03 |
| 05 | 0.996 | 0.002 | 0.002 | 0.0 |
| 06 | 0.034 | 0.54 | 0.049 | 0.377 |
| 07 | 0.191 | 0.427 | 0.065 | 0.318 |
| 08 | 0.093 | 0.105 | 0.104 | 0.697 |
| 09 | 0.115 | 0.01 | 0.831 | 0.044 |
| 10 | 0.993 | 0.0 | 0.006 | 0.001 |
| 11 | 0.064 | 0.182 | 0.276 | 0.479 |
| 12 | 0.528 | 0.399 | 0.02 | 0.053 |
| 13 | 0.082 | 0.586 | 0.056 | 0.276 |
| 14 | 0.097 | 0.409 | 0.236 | 0.257 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.194 | -2.457 | -0.185 | 0.815 |
| 02 | -3.49 | -3.982 | 1.365 | -3.369 |
| 03 | 0.699 | -1.735 | -1.735 | 0.492 |
| 04 | 1.29 | -1.589 | -3.26 | -2.078 |
| 05 | 1.377 | -4.219 | -4.219 | -4.988 |
| 06 | -1.959 | 0.767 | -1.609 | 0.409 |
| 07 | -0.269 | 0.533 | -1.335 | 0.238 |
| 08 | -0.974 | -0.856 | -0.865 | 1.021 |
| 09 | -0.767 | -3.073 | 1.197 | -1.713 |
| 10 | 1.374 | -4.988 | -3.49 | -4.531 |
| 11 | -1.35 | -0.316 | 0.099 | 0.646 |
| 12 | 0.745 | 0.465 | -2.457 | -1.533 |
| 13 | -1.096 | 0.848 | -1.48 | 0.099 |
| 14 | -0.933 | 0.49 | -0.056 | 0.028 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.92946 |
| 0.0005 | 5.01556 |
| 0.0001 | 7.194815 |