| Motif | PITX3.H14INVIVO.1.S.D |
| Gene (human) | PITX3 (GeneCards) |
| Gene synonyms (human) | PTX3 |
| Gene (mouse) | Pitx3 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 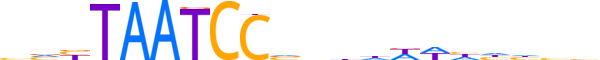 |
| Motif subtype | 1 |
| Quality | D |
| Motif | PITX3.H14INVIVO.1.S.D |
| Gene (human) | PITX3 (GeneCards) |
| Gene synonyms (human) | PTX3 |
| Gene (mouse) | Pitx3 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 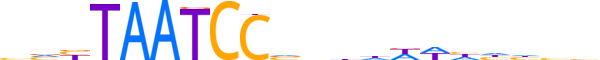 |
| Motif subtype | 1 |
| Quality | D |
| Motif length | 20 |
| Consensus | vvdddWWhvnbGGATTAvbb |
| GC content | 43.28% |
| Information content (bits; total / per base) | 14.165 / 0.708 |
| Data sources | HT-SELEX |
| Aligned words | 446 |
| Previous names | PITX3.H12INVIVO.1.S.D |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 0.976 | 0.959 | 0.955 | 0.928 | 0.872 | 0.86 |
| best | 0.994 | 0.991 | 0.977 | 0.97 | 0.943 | 0.918 | |
| Methyl HT-SELEX, 2 experiments | median | 0.97 | 0.948 | 0.956 | 0.93 | 0.921 | 0.892 |
| best | 0.977 | 0.96 | 0.968 | 0.947 | 0.943 | 0.918 | |
| Non-Methyl HT-SELEX, 4 experiments | median | 0.976 | 0.961 | 0.934 | 0.916 | 0.831 | 0.821 |
| best | 0.994 | 0.991 | 0.977 | 0.97 | 0.935 | 0.909 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.651 | 0.275 | 0.591 | 0.376 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | Paired-related HD {3.1.3} (TFClass) |
| TF subfamily | PITX {3.1.3.19} (TFClass) |
| TFClass ID | TFClass: 3.1.3.19.3 |
| HGNC | HGNC:9006 |
| MGI | MGI:1100498 |
| EntrezGene (human) | GeneID:5309 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:18742 (SSTAR profile) |
| UniProt ID (human) | PITX3_HUMAN |
| UniProt ID (mouse) | PITX3_MOUSE |
| UniProt AC (human) | O75364 (TFClass) |
| UniProt AC (mouse) | O35160 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 4 |
| Methyl-HT-SELEX | 2 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | PITX3.H14INVIVO.1.S.D.pcm |
| PWM | PITX3.H14INVIVO.1.S.D.pwm |
| PFM | PITX3.H14INVIVO.1.S.D.pfm |
| Threshold to P-value map | PITX3.H14INVIVO.1.S.D.thr |
| Motif in other formats | |
| JASPAR format | PITX3.H14INVIVO.1.S.D_jaspar_format.txt |
| MEME format | PITX3.H14INVIVO.1.S.D_meme_format.meme |
| Transfac format | PITX3.H14INVIVO.1.S.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 81.5 | 122.5 | 177.5 | 64.5 |
| 02 | 85.25 | 101.25 | 195.25 | 64.25 |
| 03 | 86.0 | 61.0 | 218.0 | 81.0 |
| 04 | 142.0 | 20.0 | 170.0 | 114.0 |
| 05 | 147.0 | 15.0 | 122.0 | 162.0 |
| 06 | 215.0 | 26.0 | 23.0 | 182.0 |
| 07 | 162.0 | 48.0 | 20.0 | 216.0 |
| 08 | 139.0 | 81.0 | 45.0 | 181.0 |
| 09 | 181.0 | 106.0 | 92.0 | 67.0 |
| 10 | 149.0 | 103.0 | 113.0 | 81.0 |
| 11 | 50.0 | 105.0 | 208.0 | 83.0 |
| 12 | 11.0 | 9.0 | 419.0 | 7.0 |
| 13 | 1.0 | 1.0 | 444.0 | 0.0 |
| 14 | 429.0 | 17.0 | 0.0 | 0.0 |
| 15 | 0.0 | 0.0 | 0.0 | 446.0 |
| 16 | 0.0 | 2.0 | 1.0 | 443.0 |
| 17 | 440.0 | 2.0 | 0.0 | 4.0 |
| 18 | 203.0 | 44.0 | 166.0 | 33.0 |
| 19 | 42.5 | 85.5 | 201.5 | 116.5 |
| 20 | 72.0 | 91.0 | 205.0 | 78.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.183 | 0.275 | 0.398 | 0.145 |
| 02 | 0.191 | 0.227 | 0.438 | 0.144 |
| 03 | 0.193 | 0.137 | 0.489 | 0.182 |
| 04 | 0.318 | 0.045 | 0.381 | 0.256 |
| 05 | 0.33 | 0.034 | 0.274 | 0.363 |
| 06 | 0.482 | 0.058 | 0.052 | 0.408 |
| 07 | 0.363 | 0.108 | 0.045 | 0.484 |
| 08 | 0.312 | 0.182 | 0.101 | 0.406 |
| 09 | 0.406 | 0.238 | 0.206 | 0.15 |
| 10 | 0.334 | 0.231 | 0.253 | 0.182 |
| 11 | 0.112 | 0.235 | 0.466 | 0.186 |
| 12 | 0.025 | 0.02 | 0.939 | 0.016 |
| 13 | 0.002 | 0.002 | 0.996 | 0.0 |
| 14 | 0.962 | 0.038 | 0.0 | 0.0 |
| 15 | 0.0 | 0.0 | 0.0 | 1.0 |
| 16 | 0.0 | 0.004 | 0.002 | 0.993 |
| 17 | 0.987 | 0.004 | 0.0 | 0.009 |
| 18 | 0.455 | 0.099 | 0.372 | 0.074 |
| 19 | 0.095 | 0.192 | 0.452 | 0.261 |
| 20 | 0.161 | 0.204 | 0.46 | 0.175 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.308 | 0.093 | 0.46 | -0.538 |
| 02 | -0.264 | -0.095 | 0.554 | -0.541 |
| 03 | -0.256 | -0.592 | 0.664 | -0.315 |
| 04 | 0.239 | -1.658 | 0.417 | 0.022 |
| 05 | 0.273 | -1.923 | 0.089 | 0.369 |
| 06 | 0.65 | -1.413 | -1.528 | 0.485 |
| 07 | 0.369 | -0.825 | -1.658 | 0.655 |
| 08 | 0.218 | -0.315 | -0.888 | 0.479 |
| 09 | 0.479 | -0.05 | -0.189 | -0.5 |
| 10 | 0.287 | -0.078 | 0.013 | -0.315 |
| 11 | -0.786 | -0.059 | 0.617 | -0.291 |
| 12 | -2.2 | -2.374 | 1.314 | -2.585 |
| 13 | -3.801 | -3.801 | 1.372 | -4.306 |
| 14 | 1.337 | -1.808 | -4.306 | -4.306 |
| 15 | -4.306 | -4.306 | -4.306 | 1.376 |
| 16 | -4.306 | -3.468 | -3.801 | 1.369 |
| 17 | 1.363 | -3.468 | -4.306 | -3.018 |
| 18 | 0.593 | -0.909 | 0.394 | -1.186 |
| 19 | -0.943 | -0.261 | 0.586 | 0.043 |
| 20 | -0.43 | -0.2 | 0.603 | -0.352 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.16231 |
| 0.0005 | 4.36321 |
| 0.0001 | 6.87746 |