| Motif | NFIA.H14RSNP.1.PS.A |
| Gene (human) | NFIA (GeneCards) |
| Gene synonyms (human) | KIAA1439 |
| Gene (mouse) | Nfia |
| Gene synonyms (mouse) | |
| LOGO | 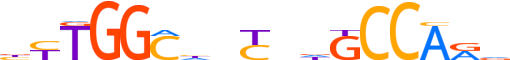 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | A |
| Motif | NFIA.H14RSNP.1.PS.A |
| Gene (human) | NFIA (GeneCards) |
| Gene synonyms (human) | KIAA1439 |
| Gene (mouse) | Nfia |
| Gene synonyms (mouse) | |
| LOGO | 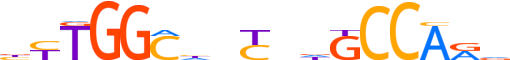 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | A |
| Motif length | 17 |
| Consensus | bYTGGChnCndGCCARv |
| GC content | 60.64% |
| Information content (bits; total / per base) | 15.419 / 0.907 |
| Data sources | ChIP-Seq + HT-SELEX |
| Aligned words | 1000 |
| Previous names | NFIA.H12RSNP.1.PS.A |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 3 (21) | 0.972 | 0.989 | 0.958 | 0.976 | 0.96 | 0.98 | 4.634 | 5.231 | 503.301 | 729.886 |
| Mouse | 6 (28) | 0.801 | 0.867 | 0.705 | 0.805 | 0.753 | 0.832 | 2.209 | 2.856 | 67.474 | 164.027 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 1.0 | 1.0 | 0.986 | 0.987 | 0.819 | 0.854 |
| best | 1.0 | 1.0 | 1.0 | 1.0 | 0.887 | 0.909 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 100.0 | 4.148 | 0.341 | 0.162 |
| TF superclass | beta-Hairpin exposed by an alpha/beta-scaffold {7} (TFClass) |
| TF class | SMAD/NF-1 DNA-binding domain factors {7.1} (TFClass) |
| TF family | NF-1 {7.1.2} (TFClass) |
| TF subfamily | {7.1.2.0} (TFClass) |
| TFClass ID | TFClass: 7.1.2.0.1 |
| HGNC | HGNC:7784 |
| MGI | MGI:108056 |
| EntrezGene (human) | GeneID:4774 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:18027 (SSTAR profile) |
| UniProt ID (human) | NFIA_HUMAN |
| UniProt ID (mouse) | NFIA_MOUSE |
| UniProt AC (human) | Q12857 (TFClass) |
| UniProt AC (mouse) | Q02780 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 3 human, 6 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | NFIA.H14RSNP.1.PS.A.pcm |
| PWM | NFIA.H14RSNP.1.PS.A.pwm |
| PFM | NFIA.H14RSNP.1.PS.A.pfm |
| Threshold to P-value map | NFIA.H14RSNP.1.PS.A.thr |
| Motif in other formats | |
| JASPAR format | NFIA.H14RSNP.1.PS.A_jaspar_format.txt |
| MEME format | NFIA.H14RSNP.1.PS.A_meme_format.meme |
| Transfac format | NFIA.H14RSNP.1.PS.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 88.0 | 458.0 | 205.0 | 249.0 |
| 02 | 27.0 | 429.0 | 33.0 | 511.0 |
| 03 | 26.0 | 2.0 | 108.0 | 864.0 |
| 04 | 0.0 | 0.0 | 997.0 | 3.0 |
| 05 | 10.0 | 4.0 | 982.0 | 4.0 |
| 06 | 192.0 | 767.0 | 29.0 | 12.0 |
| 07 | 388.0 | 235.0 | 70.0 | 307.0 |
| 08 | 159.0 | 386.0 | 236.0 | 219.0 |
| 09 | 0.0 | 506.0 | 0.0 | 494.0 |
| 10 | 173.0 | 304.0 | 287.0 | 236.0 |
| 11 | 250.0 | 78.0 | 230.0 | 442.0 |
| 12 | 10.0 | 22.0 | 764.0 | 204.0 |
| 13 | 5.0 | 980.0 | 0.0 | 15.0 |
| 14 | 2.0 | 993.0 | 2.0 | 3.0 |
| 15 | 864.0 | 105.0 | 6.0 | 25.0 |
| 16 | 506.0 | 35.0 | 414.0 | 45.0 |
| 17 | 248.0 | 209.0 | 432.0 | 111.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.088 | 0.458 | 0.205 | 0.249 |
| 02 | 0.027 | 0.429 | 0.033 | 0.511 |
| 03 | 0.026 | 0.002 | 0.108 | 0.864 |
| 04 | 0.0 | 0.0 | 0.997 | 0.003 |
| 05 | 0.01 | 0.004 | 0.982 | 0.004 |
| 06 | 0.192 | 0.767 | 0.029 | 0.012 |
| 07 | 0.388 | 0.235 | 0.07 | 0.307 |
| 08 | 0.159 | 0.386 | 0.236 | 0.219 |
| 09 | 0.0 | 0.506 | 0.0 | 0.494 |
| 10 | 0.173 | 0.304 | 0.287 | 0.236 |
| 11 | 0.25 | 0.078 | 0.23 | 0.442 |
| 12 | 0.01 | 0.022 | 0.764 | 0.204 |
| 13 | 0.005 | 0.98 | 0.0 | 0.015 |
| 14 | 0.002 | 0.993 | 0.002 | 0.003 |
| 15 | 0.864 | 0.105 | 0.006 | 0.025 |
| 16 | 0.506 | 0.035 | 0.414 | 0.045 |
| 17 | 0.248 | 0.209 | 0.432 | 0.111 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -1.032 | 0.602 | -0.197 | -0.004 |
| 02 | -2.171 | 0.537 | -1.981 | 0.711 |
| 03 | -2.206 | -4.213 | -0.83 | 1.235 |
| 04 | -4.982 | -4.982 | 1.378 | -3.975 |
| 05 | -3.066 | -3.783 | 1.363 | -3.783 |
| 06 | -0.262 | 1.116 | -2.103 | -2.909 |
| 07 | 0.437 | -0.061 | -1.255 | 0.204 |
| 08 | -0.449 | 0.432 | -0.057 | -0.131 |
| 09 | -4.982 | 0.702 | -4.982 | 0.678 |
| 10 | -0.365 | 0.194 | 0.137 | -0.057 |
| 11 | 0.0 | -1.15 | -0.083 | 0.567 |
| 12 | -3.066 | -2.362 | 1.112 | -0.202 |
| 13 | -3.622 | 1.361 | -4.982 | -2.711 |
| 14 | -4.213 | 1.374 | -4.213 | -3.975 |
| 15 | 1.235 | -0.858 | -3.484 | -2.243 |
| 16 | 0.702 | -1.925 | 0.502 | -1.684 |
| 17 | -0.008 | -0.178 | 0.544 | -0.803 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.28021 |
| 0.0005 | 3.61316 |
| 0.0001 | 6.37296 |