| Motif | NFAC2.H14CORE.0.P.B |
| Gene (human) | NFATC2 (GeneCards) |
| Gene synonyms (human) | NFAT1, NFATP |
| Gene (mouse) | Nfatc2 |
| Gene synonyms (mouse) | Nfat1, Nfatp |
| LOGO |  |
| LOGO (reverse complement) | 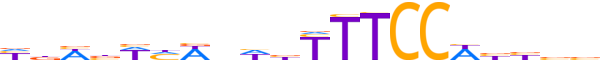 |
| Motif subtype | 0 |
| Quality | B |
| Motif | NFAC2.H14CORE.0.P.B |
| Gene (human) | NFATC2 (GeneCards) |
| Gene synonyms (human) | NFAT1, NFATP |
| Gene (mouse) | Nfatc2 |
| Gene synonyms (mouse) | Nfat1, Nfatp |
| LOGO |  |
| LOGO (reverse complement) | 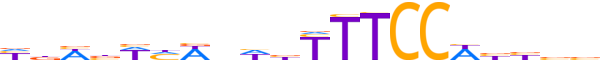 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 20 |
| Consensus | vvvvWGGAARvdnWdWvWvd |
| GC content | 41.52% |
| Information content (bits; total / per base) | 12.814 / 0.641 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| Previous names | NFAC2.H12CORE.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (10) | 0.84 | 0.879 | 0.751 | 0.812 | 0.889 | 0.9 | 3.649 | 3.843 | 157.327 | 243.959 |
| Mouse | 1 (7) | 0.859 | 0.87 | 0.741 | 0.76 | 0.855 | 0.859 | 2.886 | 2.908 | 160.886 | 178.678 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 4 experiments | median | 0.945 | 0.912 | 0.921 | 0.883 | 0.876 | 0.837 |
| best | 0.98 | 0.967 | 0.97 | 0.952 | 0.944 | 0.921 | |
| Methyl HT-SELEX, 2 experiments | median | 0.927 | 0.886 | 0.9 | 0.856 | 0.852 | 0.811 |
| best | 0.966 | 0.942 | 0.948 | 0.919 | 0.907 | 0.875 | |
| Non-Methyl HT-SELEX, 2 experiments | median | 0.952 | 0.924 | 0.932 | 0.899 | 0.894 | 0.859 |
| best | 0.98 | 0.967 | 0.97 | 0.952 | 0.944 | 0.921 | |
| TF superclass | Immunoglobulin fold {6} (TFClass) |
| TF class | Rel homology region (RHR) factors {6.1} (TFClass) |
| TF family | NFAT-related {6.1.3} (TFClass) |
| TF subfamily | {6.1.3.0} (TFClass) |
| TFClass ID | TFClass: 6.1.3.0.2 |
| HGNC | HGNC:7776 |
| MGI | MGI:102463 |
| EntrezGene (human) | GeneID:4773 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:18019 (SSTAR profile) |
| UniProt ID (human) | NFAC2_HUMAN |
| UniProt ID (mouse) | NFAC2_MOUSE |
| UniProt AC (human) | Q13469 (TFClass) |
| UniProt AC (mouse) | Q60591 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 1 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 2 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | NFAC2.H14CORE.0.P.B.pcm |
| PWM | NFAC2.H14CORE.0.P.B.pwm |
| PFM | NFAC2.H14CORE.0.P.B.pfm |
| Threshold to P-value map | NFAC2.H14CORE.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | NFAC2.H14CORE.0.P.B_jaspar_format.txt |
| MEME format | NFAC2.H14CORE.0.P.B_meme_format.meme |
| Transfac format | NFAC2.H14CORE.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 180.0 | 268.0 | 416.0 | 136.0 |
| 02 | 174.0 | 266.0 | 424.0 | 136.0 |
| 03 | 525.0 | 137.0 | 251.0 | 87.0 |
| 04 | 483.0 | 135.0 | 283.0 | 99.0 |
| 05 | 240.0 | 53.0 | 71.0 | 636.0 |
| 06 | 8.0 | 0.0 | 992.0 | 0.0 |
| 07 | 2.0 | 4.0 | 994.0 | 0.0 |
| 08 | 949.0 | 20.0 | 26.0 | 5.0 |
| 09 | 927.0 | 7.0 | 58.0 | 8.0 |
| 10 | 797.0 | 45.0 | 96.0 | 62.0 |
| 11 | 499.0 | 157.0 | 198.0 | 146.0 |
| 12 | 478.0 | 46.0 | 115.0 | 361.0 |
| 13 | 379.0 | 187.0 | 218.0 | 216.0 |
| 14 | 166.0 | 119.0 | 61.0 | 654.0 |
| 15 | 149.0 | 83.0 | 567.0 | 201.0 |
| 16 | 648.0 | 77.0 | 110.0 | 165.0 |
| 17 | 201.0 | 300.0 | 411.0 | 88.0 |
| 18 | 165.0 | 98.0 | 112.0 | 625.0 |
| 19 | 226.0 | 488.0 | 150.0 | 136.0 |
| 20 | 589.0 | 106.0 | 155.0 | 150.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.18 | 0.268 | 0.416 | 0.136 |
| 02 | 0.174 | 0.266 | 0.424 | 0.136 |
| 03 | 0.525 | 0.137 | 0.251 | 0.087 |
| 04 | 0.483 | 0.135 | 0.283 | 0.099 |
| 05 | 0.24 | 0.053 | 0.071 | 0.636 |
| 06 | 0.008 | 0.0 | 0.992 | 0.0 |
| 07 | 0.002 | 0.004 | 0.994 | 0.0 |
| 08 | 0.949 | 0.02 | 0.026 | 0.005 |
| 09 | 0.927 | 0.007 | 0.058 | 0.008 |
| 10 | 0.797 | 0.045 | 0.096 | 0.062 |
| 11 | 0.499 | 0.157 | 0.198 | 0.146 |
| 12 | 0.478 | 0.046 | 0.115 | 0.361 |
| 13 | 0.379 | 0.187 | 0.218 | 0.216 |
| 14 | 0.166 | 0.119 | 0.061 | 0.654 |
| 15 | 0.149 | 0.083 | 0.567 | 0.201 |
| 16 | 0.648 | 0.077 | 0.11 | 0.165 |
| 17 | 0.201 | 0.3 | 0.411 | 0.088 |
| 18 | 0.165 | 0.098 | 0.112 | 0.625 |
| 19 | 0.226 | 0.488 | 0.15 | 0.136 |
| 20 | 0.589 | 0.106 | 0.155 | 0.15 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.326 | 0.069 | 0.506 | -0.603 |
| 02 | -0.359 | 0.062 | 0.525 | -0.603 |
| 03 | 0.738 | -0.596 | 0.004 | -1.043 |
| 04 | 0.655 | -0.61 | 0.123 | -0.916 |
| 05 | -0.041 | -1.526 | -1.242 | 0.93 |
| 06 | -3.253 | -4.982 | 1.373 | -4.982 |
| 07 | -4.213 | -3.783 | 1.375 | -4.982 |
| 08 | 1.329 | -2.45 | -2.206 | -3.622 |
| 09 | 1.305 | -3.362 | -1.439 | -3.253 |
| 10 | 1.155 | -1.684 | -0.946 | -1.374 |
| 11 | 0.688 | -0.461 | -0.231 | -0.533 |
| 12 | 0.645 | -1.663 | -0.769 | 0.365 |
| 13 | 0.414 | -0.288 | -0.136 | -0.145 |
| 14 | -0.406 | -0.735 | -1.39 | 0.957 |
| 15 | -0.513 | -1.089 | 0.815 | -0.216 |
| 16 | 0.948 | -1.162 | -0.812 | -0.412 |
| 17 | -0.216 | 0.181 | 0.494 | -1.032 |
| 18 | -0.412 | -0.926 | -0.795 | 0.912 |
| 19 | -0.1 | 0.666 | -0.506 | -0.603 |
| 20 | 0.853 | -0.849 | -0.474 | -0.506 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.99546 |
| 0.0005 | 5.01306 |
| 0.0001 | 7.07596 |