| Motif | MSX1.H14RSNP.1.S.C |
| Gene (human) | MSX1 (GeneCards) |
| Gene synonyms (human) | HOX7 |
| Gene (mouse) | Msx1 |
| Gene synonyms (mouse) | Hox7, Hox7.1 |
| LOGO |  |
| LOGO (reverse complement) | 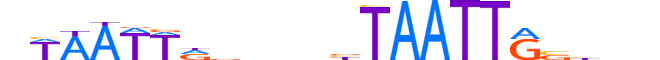 |
| Motif subtype | 1 |
| Quality | C |
| Motif | MSX1.H14RSNP.1.S.C |
| Gene (human) | MSX1 (GeneCards) |
| Gene synonyms (human) | HOX7 |
| Gene (mouse) | Msx1 |
| Gene synonyms (mouse) | Hox7, Hox7.1 |
| LOGO |  |
| LOGO (reverse complement) | 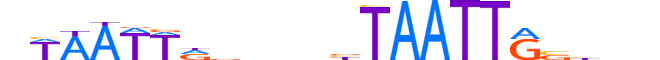 |
| Motif subtype | 1 |
| Quality | C |
| Motif length | 22 |
| Consensus | nnvbCAATTAvnnnbhAATTWn |
| GC content | 31.04% |
| Information content (bits; total / per base) | 17.53 / 0.797 |
| Data sources | HT-SELEX |
| Aligned words | 4727 |
| Previous names | MSX1.H12RSNP.1.S.C |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 8 experiments | median | 0.869 | 0.836 | 0.729 | 0.718 | 0.622 | 0.634 |
| best | 0.995 | 0.993 | 0.934 | 0.93 | 0.812 | 0.783 | |
| Methyl HT-SELEX, 2 experiments | median | 0.886 | 0.848 | 0.798 | 0.773 | 0.704 | 0.697 |
| best | 0.935 | 0.895 | 0.902 | 0.858 | 0.81 | 0.782 | |
| Non-Methyl HT-SELEX, 6 experiments | median | 0.869 | 0.836 | 0.729 | 0.718 | 0.622 | 0.634 |
| best | 0.995 | 0.993 | 0.934 | 0.93 | 0.812 | 0.783 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.765 | 0.441 | 0.734 | 0.561 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | NK-related {3.1.2} (TFClass) |
| TF subfamily | MSX {3.1.2.11} (TFClass) |
| TFClass ID | TFClass: 3.1.2.11.1 |
| HGNC | HGNC:7391 |
| MGI | MGI:97168 |
| EntrezGene (human) | GeneID:4487 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:17701 (SSTAR profile) |
| UniProt ID (human) | MSX1_HUMAN |
| UniProt ID (mouse) | MSX1_MOUSE |
| UniProt AC (human) | P28360 (TFClass) |
| UniProt AC (mouse) | P13297 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 6 |
| Methyl-HT-SELEX | 2 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | MSX1.H14RSNP.1.S.C.pcm |
| PWM | MSX1.H14RSNP.1.S.C.pwm |
| PFM | MSX1.H14RSNP.1.S.C.pfm |
| Threshold to P-value map | MSX1.H14RSNP.1.S.C.thr |
| Motif in other formats | |
| JASPAR format | MSX1.H14RSNP.1.S.C_jaspar_format.txt |
| MEME format | MSX1.H14RSNP.1.S.C_meme_format.meme |
| Transfac format | MSX1.H14RSNP.1.S.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1625.75 | 762.75 | 1065.75 | 1272.75 |
| 02 | 1415.5 | 1055.5 | 1565.5 | 690.5 |
| 03 | 2022.0 | 820.0 | 1297.0 | 588.0 |
| 04 | 382.0 | 2165.0 | 1745.0 | 435.0 |
| 05 | 4.0 | 3111.0 | 7.0 | 1605.0 |
| 06 | 4713.0 | 5.0 | 9.0 | 0.0 |
| 07 | 4727.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 4727.0 |
| 09 | 0.0 | 8.0 | 0.0 | 4719.0 |
| 10 | 4591.0 | 0.0 | 134.0 | 2.0 |
| 11 | 1933.0 | 801.0 | 1615.0 | 378.0 |
| 12 | 1248.0 | 1549.0 | 934.0 | 996.0 |
| 13 | 1524.0 | 818.0 | 1217.0 | 1168.0 |
| 14 | 1807.0 | 1089.0 | 880.0 | 951.0 |
| 15 | 784.0 | 1950.0 | 1147.0 | 846.0 |
| 16 | 657.0 | 2355.0 | 180.0 | 1535.0 |
| 17 | 3833.0 | 273.0 | 211.0 | 410.0 |
| 18 | 3830.0 | 155.0 | 85.0 | 657.0 |
| 19 | 447.0 | 105.0 | 68.0 | 4107.0 |
| 20 | 721.0 | 127.0 | 139.0 | 3740.0 |
| 21 | 3527.0 | 187.0 | 468.0 | 545.0 |
| 22 | 1565.75 | 954.75 | 1224.75 | 981.75 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.344 | 0.161 | 0.225 | 0.269 |
| 02 | 0.299 | 0.223 | 0.331 | 0.146 |
| 03 | 0.428 | 0.173 | 0.274 | 0.124 |
| 04 | 0.081 | 0.458 | 0.369 | 0.092 |
| 05 | 0.001 | 0.658 | 0.001 | 0.34 |
| 06 | 0.997 | 0.001 | 0.002 | 0.0 |
| 07 | 1.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 1.0 |
| 09 | 0.0 | 0.002 | 0.0 | 0.998 |
| 10 | 0.971 | 0.0 | 0.028 | 0.0 |
| 11 | 0.409 | 0.169 | 0.342 | 0.08 |
| 12 | 0.264 | 0.328 | 0.198 | 0.211 |
| 13 | 0.322 | 0.173 | 0.257 | 0.247 |
| 14 | 0.382 | 0.23 | 0.186 | 0.201 |
| 15 | 0.166 | 0.413 | 0.243 | 0.179 |
| 16 | 0.139 | 0.498 | 0.038 | 0.325 |
| 17 | 0.811 | 0.058 | 0.045 | 0.087 |
| 18 | 0.81 | 0.033 | 0.018 | 0.139 |
| 19 | 0.095 | 0.022 | 0.014 | 0.869 |
| 20 | 0.153 | 0.027 | 0.029 | 0.791 |
| 21 | 0.746 | 0.04 | 0.099 | 0.115 |
| 22 | 0.331 | 0.202 | 0.259 | 0.208 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.318 | -0.437 | -0.103 | 0.074 |
| 02 | 0.18 | -0.113 | 0.281 | -0.536 |
| 03 | 0.536 | -0.365 | 0.093 | -0.696 |
| 04 | -1.126 | 0.605 | 0.389 | -0.996 |
| 05 | -5.266 | 0.967 | -4.867 | 0.306 |
| 06 | 1.382 | -5.114 | -4.668 | -6.327 |
| 07 | 1.385 | -6.327 | -6.327 | -6.327 |
| 08 | -6.327 | -6.327 | -6.327 | 1.385 |
| 09 | -6.327 | -4.762 | -6.327 | 1.383 |
| 10 | 1.356 | -6.327 | -2.163 | -5.662 |
| 11 | 0.491 | -0.388 | 0.312 | -1.136 |
| 12 | 0.054 | 0.27 | -0.235 | -0.171 |
| 13 | 0.254 | -0.367 | 0.029 | -0.012 |
| 14 | 0.424 | -0.082 | -0.294 | -0.217 |
| 15 | -0.409 | 0.5 | -0.03 | -0.334 |
| 16 | -0.586 | 0.689 | -1.872 | 0.261 |
| 17 | 1.175 | -1.459 | -1.715 | -1.055 |
| 18 | 1.175 | -2.02 | -2.609 | -0.586 |
| 19 | -0.969 | -2.403 | -2.826 | 1.244 |
| 20 | -0.493 | -2.216 | -2.127 | 1.151 |
| 21 | 1.092 | -1.834 | -0.924 | -0.772 |
| 22 | 0.281 | -0.213 | 0.036 | -0.185 |
| P-value | Threshold |
|---|---|
| 0.001 | 0.05931 |
| 0.0005 | 1.75236 |
| 0.0001 | 5.27306 |