| Motif | MAFB.H14CORE.1.P.B |
| Gene (human) | MAFB (GeneCards) |
| Gene synonyms (human) | KRML |
| Gene (mouse) | Mafb |
| Gene synonyms (mouse) | Krml, Maf1 |
| LOGO |  |
| LOGO (reverse complement) | 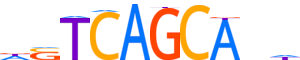 |
| Motif subtype | 1 |
| Quality | B |
| Motif | MAFB.H14CORE.1.P.B |
| Gene (human) | MAFB (GeneCards) |
| Gene synonyms (human) | KRML |
| Gene (mouse) | Mafb |
| Gene synonyms (mouse) | Krml, Maf1 |
| LOGO |  |
| LOGO (reverse complement) | 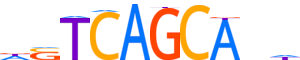 |
| Motif subtype | 1 |
| Quality | B |
| Motif length | 10 |
| Consensus | hnTGCTGASh |
| GC content | 49.04% |
| Information content (bits; total / per base) | 11.226 / 1.123 |
| Data sources | ChIP-Seq |
| Aligned words | 1001 |
| Previous names | MAFB.H12CORE.1.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 3 (16) | 0.864 | 0.883 | 0.761 | 0.789 | 0.829 | 0.847 | 2.628 | 2.787 | 250.014 | 396.0 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.929 | 0.919 | 0.848 | 0.847 | 0.747 | 0.76 |
| best | 0.996 | 0.994 | 0.991 | 0.987 | 0.886 | 0.895 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.542 | 0.013 | 0.307 | 0.26 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic leucine zipper factors (bZIP) {1.1} (TFClass) |
| TF family | Maf-related {1.1.3} (TFClass) |
| TF subfamily | Large MAF {1.1.3.1} (TFClass) |
| TFClass ID | TFClass: 1.1.3.1.3 |
| HGNC | HGNC:6408 |
| MGI | MGI:104555 |
| EntrezGene (human) | GeneID:9935 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:16658 (SSTAR profile) |
| UniProt ID (human) | MAFB_HUMAN |
| UniProt ID (mouse) | MAFB_MOUSE |
| UniProt AC (human) | Q9Y5Q3 (TFClass) |
| UniProt AC (mouse) | P54841 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 3 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | MAFB.H14CORE.1.P.B.pcm |
| PWM | MAFB.H14CORE.1.P.B.pwm |
| PFM | MAFB.H14CORE.1.P.B.pfm |
| Threshold to P-value map | MAFB.H14CORE.1.P.B.thr |
| Motif in other formats | |
| JASPAR format | MAFB.H14CORE.1.P.B_jaspar_format.txt |
| MEME format | MAFB.H14CORE.1.P.B_meme_format.meme |
| Transfac format | MAFB.H14CORE.1.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 448.0 | 115.0 | 109.0 | 329.0 |
| 02 | 181.0 | 352.0 | 213.0 | 255.0 |
| 03 | 54.0 | 26.0 | 7.0 | 914.0 |
| 04 | 0.0 | 11.0 | 985.0 | 5.0 |
| 05 | 25.0 | 972.0 | 4.0 | 0.0 |
| 06 | 13.0 | 0.0 | 0.0 | 988.0 |
| 07 | 3.0 | 8.0 | 950.0 | 40.0 |
| 08 | 926.0 | 10.0 | 18.0 | 47.0 |
| 09 | 48.0 | 561.0 | 298.0 | 94.0 |
| 10 | 229.0 | 167.0 | 103.0 | 502.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.448 | 0.115 | 0.109 | 0.329 |
| 02 | 0.181 | 0.352 | 0.213 | 0.255 |
| 03 | 0.054 | 0.026 | 0.007 | 0.913 |
| 04 | 0.0 | 0.011 | 0.984 | 0.005 |
| 05 | 0.025 | 0.971 | 0.004 | 0.0 |
| 06 | 0.013 | 0.0 | 0.0 | 0.987 |
| 07 | 0.003 | 0.008 | 0.949 | 0.04 |
| 08 | 0.925 | 0.01 | 0.018 | 0.047 |
| 09 | 0.048 | 0.56 | 0.298 | 0.094 |
| 10 | 0.229 | 0.167 | 0.103 | 0.501 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.579 | -0.769 | -0.822 | 0.272 |
| 02 | -0.321 | 0.339 | -0.16 | 0.019 |
| 03 | -1.509 | -2.207 | -3.363 | 1.29 |
| 04 | -4.983 | -2.986 | 1.365 | -3.623 |
| 05 | -2.244 | 1.352 | -3.784 | -4.983 |
| 06 | -2.84 | -4.983 | -4.983 | 1.368 |
| 07 | -3.976 | -3.254 | 1.329 | -1.798 |
| 08 | 1.303 | -3.067 | -2.547 | -1.643 |
| 09 | -1.623 | 0.803 | 0.174 | -0.968 |
| 10 | -0.088 | -0.401 | -0.878 | 0.693 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.343365 |
| 0.0005 | 5.27089 |
| 0.0001 | 7.563255 |