| Motif | LHX2.H14INVIVO.1.S.C |
| Gene (human) | LHX2 (GeneCards) |
| Gene synonyms (human) | LH2 |
| Gene (mouse) | Lhx2 |
| Gene synonyms (mouse) | |
| LOGO | 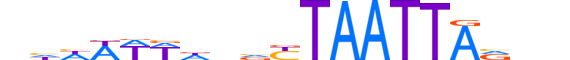 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | C |
| Motif | LHX2.H14INVIVO.1.S.C |
| Gene (human) | LHX2 (GeneCards) |
| Gene synonyms (human) | LH2 |
| Gene (mouse) | Lhx2 |
| Gene synonyms (mouse) | |
| LOGO | 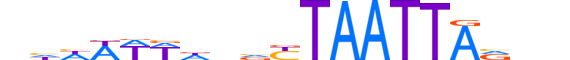 |
| LOGO (reverse complement) |  |
| Motif subtype | 1 |
| Quality | C |
| Motif length | 19 |
| Consensus | nhhWWWdnvYTAATTAvnn |
| GC content | 26.26% |
| Information content (bits; total / per base) | 15.479 / 0.815 |
| Data sources | HT-SELEX |
| Aligned words | 2014 |
| Previous names | LHX2.H12INVIVO.1.S.C |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (6) | 0.786 | 0.795 | 0.576 | 0.581 | 0.807 | 0.821 | 2.36 | 2.385 | 89.699 | 97.018 |
| Mouse | 9 (53) | 0.851 | 0.922 | 0.677 | 0.822 | 0.777 | 0.883 | 2.558 | 3.172 | 126.222 | 203.678 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.769 | 0.746 | 0.64 | 0.643 | 0.569 | 0.584 |
| best | 0.867 | 0.849 | 0.693 | 0.704 | 0.592 | 0.619 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | HD-LIM {3.1.5} (TFClass) |
| TF subfamily | LHX2-like {3.1.5.3} (TFClass) |
| TFClass ID | TFClass: 3.1.5.3.1 |
| HGNC | HGNC:6594 |
| MGI | MGI:96785 |
| EntrezGene (human) | GeneID:9355 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:16870 (SSTAR profile) |
| UniProt ID (human) | LHX2_HUMAN |
| UniProt ID (mouse) | LHX2_MOUSE |
| UniProt AC (human) | P50458 (TFClass) |
| UniProt AC (mouse) | Q9Z0S2 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 9 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | LHX2.H14INVIVO.1.S.C.pcm |
| PWM | LHX2.H14INVIVO.1.S.C.pwm |
| PFM | LHX2.H14INVIVO.1.S.C.pfm |
| Threshold to P-value map | LHX2.H14INVIVO.1.S.C.thr |
| Motif in other formats | |
| JASPAR format | LHX2.H14INVIVO.1.S.C_jaspar_format.txt |
| MEME format | LHX2.H14INVIVO.1.S.C_meme_format.meme |
| Transfac format | LHX2.H14INVIVO.1.S.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 569.75 | 461.75 | 291.75 | 690.75 |
| 02 | 517.0 | 492.0 | 121.0 | 884.0 |
| 03 | 1097.0 | 314.0 | 104.0 | 499.0 |
| 04 | 1351.0 | 81.0 | 121.0 | 461.0 |
| 05 | 553.0 | 59.0 | 103.0 | 1299.0 |
| 06 | 391.0 | 110.0 | 167.0 | 1346.0 |
| 07 | 1051.0 | 161.0 | 285.0 | 517.0 |
| 08 | 711.0 | 439.0 | 502.0 | 362.0 |
| 09 | 577.0 | 387.0 | 873.0 | 177.0 |
| 10 | 87.0 | 1193.0 | 200.0 | 534.0 |
| 11 | 0.0 | 2.0 | 0.0 | 2012.0 |
| 12 | 2010.0 | 4.0 | 0.0 | 0.0 |
| 13 | 2014.0 | 0.0 | 0.0 | 0.0 |
| 14 | 0.0 | 0.0 | 0.0 | 2014.0 |
| 15 | 1.0 | 1.0 | 20.0 | 1992.0 |
| 16 | 1697.0 | 1.0 | 315.0 | 1.0 |
| 17 | 607.0 | 282.0 | 1031.0 | 94.0 |
| 18 | 304.75 | 565.75 | 483.75 | 659.75 |
| 19 | 552.5 | 387.5 | 488.5 | 585.5 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.283 | 0.229 | 0.145 | 0.343 |
| 02 | 0.257 | 0.244 | 0.06 | 0.439 |
| 03 | 0.545 | 0.156 | 0.052 | 0.248 |
| 04 | 0.671 | 0.04 | 0.06 | 0.229 |
| 05 | 0.275 | 0.029 | 0.051 | 0.645 |
| 06 | 0.194 | 0.055 | 0.083 | 0.668 |
| 07 | 0.522 | 0.08 | 0.142 | 0.257 |
| 08 | 0.353 | 0.218 | 0.249 | 0.18 |
| 09 | 0.286 | 0.192 | 0.433 | 0.088 |
| 10 | 0.043 | 0.592 | 0.099 | 0.265 |
| 11 | 0.0 | 0.001 | 0.0 | 0.999 |
| 12 | 0.998 | 0.002 | 0.0 | 0.0 |
| 13 | 1.0 | 0.0 | 0.0 | 0.0 |
| 14 | 0.0 | 0.0 | 0.0 | 1.0 |
| 15 | 0.0 | 0.0 | 0.01 | 0.989 |
| 16 | 0.843 | 0.0 | 0.156 | 0.0 |
| 17 | 0.301 | 0.14 | 0.512 | 0.047 |
| 18 | 0.151 | 0.281 | 0.24 | 0.328 |
| 19 | 0.274 | 0.192 | 0.243 | 0.291 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.123 | -0.086 | -0.543 | 0.315 |
| 02 | 0.026 | -0.023 | -1.414 | 0.561 |
| 03 | 0.777 | -0.47 | -1.563 | -0.009 |
| 04 | 0.985 | -1.808 | -1.414 | -0.088 |
| 05 | 0.093 | -2.116 | -1.572 | 0.945 |
| 06 | -0.252 | -1.508 | -1.096 | 0.981 |
| 07 | 0.734 | -1.132 | -0.566 | 0.026 |
| 08 | 0.344 | -0.137 | -0.003 | -0.328 |
| 09 | 0.136 | -0.262 | 0.549 | -1.039 |
| 10 | -1.738 | 0.86 | -0.918 | 0.059 |
| 11 | -5.582 | -4.864 | -5.582 | 1.382 |
| 12 | 1.381 | -4.45 | -5.582 | -5.582 |
| 13 | 1.383 | -5.582 | -5.582 | -5.582 |
| 14 | -5.582 | -5.582 | -5.582 | 1.383 |
| 15 | -5.16 | -5.16 | -3.139 | 1.372 |
| 16 | 1.212 | -5.16 | -0.467 | -5.16 |
| 17 | 0.186 | -0.577 | 0.715 | -1.662 |
| 18 | -0.5 | 0.116 | -0.04 | 0.269 |
| 19 | 0.093 | -0.261 | -0.03 | 0.15 |
| P-value | Threshold |
|---|---|
| 0.001 | 1.46291 |
| 0.0005 | 2.97301 |
| 0.0001 | 6.32036 |