| Motif | KLF10.H14INVIVO.0.P.C |
| Gene (human) | KLF10 (GeneCards) |
| Gene synonyms (human) | TIEG, TIEG1 |
| Gene (mouse) | Klf10 |
| Gene synonyms (mouse) | Gdnfif, Tieg, Tieg1 |
| LOGO | 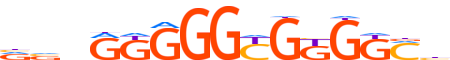 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | C |
| Motif | KLF10.H14INVIVO.0.P.C |
| Gene (human) | KLF10 (GeneCards) |
| Gene synonyms (human) | TIEG, TIEG1 |
| Gene (mouse) | Klf10 |
| Gene synonyms (mouse) | Gdnfif, Tieg, Tieg1 |
| LOGO | 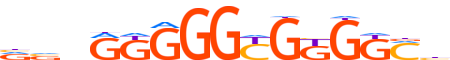 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | C |
| Motif length | 15 |
| Consensus | vvnRKGGGYGKGKMb |
| GC content | 77.87% |
| Information content (bits; total / per base) | 13.776 / 0.918 |
| Data sources | ChIP-Seq |
| Aligned words | 978 |
| Previous names | KLF10.H12INVIVO.0.P.C |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 1 (7) | 0.801 | 0.853 | 0.662 | 0.689 | 0.846 | 0.91 | 3.271 | 3.484 | 44.367 | 52.187 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.914 | 0.861 | 0.877 | 0.825 | 0.781 | 0.754 |
| best | 0.923 | 0.872 | 0.896 | 0.844 | 0.838 | 0.793 | |
| Methyl HT-SELEX, 1 experiments | median | 0.923 | 0.872 | 0.896 | 0.844 | 0.838 | 0.793 |
| best | 0.923 | 0.872 | 0.896 | 0.844 | 0.838 | 0.793 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.905 | 0.85 | 0.859 | 0.806 | 0.724 | 0.716 |
| best | 0.905 | 0.85 | 0.859 | 0.806 | 0.724 | 0.716 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.899 | 0.049 | 0.784 | 0.5 |
| batch 2 | 0.525 | 0.124 | 0.542 | 0.339 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | Three-zinc finger Kruppel-related {2.3.1} (TFClass) |
| TF subfamily | Kr-like {2.3.1.2} (TFClass) |
| TFClass ID | TFClass: 2.3.1.2.10 |
| HGNC | HGNC:11810 |
| MGI | MGI:1101353 |
| EntrezGene (human) | GeneID:7071 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:21847 (SSTAR profile) |
| UniProt ID (human) | KLF10_HUMAN |
| UniProt ID (mouse) | KLF10_MOUSE |
| UniProt AC (human) | Q13118 (TFClass) |
| UniProt AC (mouse) | O89091 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 1 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | KLF10.H14INVIVO.0.P.C.pcm |
| PWM | KLF10.H14INVIVO.0.P.C.pwm |
| PFM | KLF10.H14INVIVO.0.P.C.pfm |
| Threshold to P-value map | KLF10.H14INVIVO.0.P.C.thr |
| Motif in other formats | |
| JASPAR format | KLF10.H14INVIVO.0.P.C_jaspar_format.txt |
| MEME format | KLF10.H14INVIVO.0.P.C_meme_format.meme |
| Transfac format | KLF10.H14INVIVO.0.P.C_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 163.0 | 162.0 | 555.0 | 98.0 |
| 02 | 195.0 | 213.0 | 482.0 | 88.0 |
| 03 | 366.0 | 206.0 | 233.0 | 173.0 |
| 04 | 173.0 | 20.0 | 747.0 | 38.0 |
| 05 | 98.0 | 12.0 | 736.0 | 132.0 |
| 06 | 155.0 | 7.0 | 812.0 | 4.0 |
| 07 | 8.0 | 8.0 | 953.0 | 9.0 |
| 08 | 6.0 | 8.0 | 960.0 | 4.0 |
| 09 | 60.0 | 650.0 | 4.0 | 264.0 |
| 10 | 23.0 | 17.0 | 908.0 | 30.0 |
| 11 | 7.0 | 54.0 | 595.0 | 322.0 |
| 12 | 39.0 | 6.0 | 890.0 | 43.0 |
| 13 | 63.0 | 25.0 | 746.0 | 144.0 |
| 14 | 101.0 | 725.0 | 90.0 | 62.0 |
| 15 | 123.0 | 431.0 | 169.0 | 255.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.167 | 0.166 | 0.567 | 0.1 |
| 02 | 0.199 | 0.218 | 0.493 | 0.09 |
| 03 | 0.374 | 0.211 | 0.238 | 0.177 |
| 04 | 0.177 | 0.02 | 0.764 | 0.039 |
| 05 | 0.1 | 0.012 | 0.753 | 0.135 |
| 06 | 0.158 | 0.007 | 0.83 | 0.004 |
| 07 | 0.008 | 0.008 | 0.974 | 0.009 |
| 08 | 0.006 | 0.008 | 0.982 | 0.004 |
| 09 | 0.061 | 0.665 | 0.004 | 0.27 |
| 10 | 0.024 | 0.017 | 0.928 | 0.031 |
| 11 | 0.007 | 0.055 | 0.608 | 0.329 |
| 12 | 0.04 | 0.006 | 0.91 | 0.044 |
| 13 | 0.064 | 0.026 | 0.763 | 0.147 |
| 14 | 0.103 | 0.741 | 0.092 | 0.063 |
| 15 | 0.126 | 0.441 | 0.173 | 0.261 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.402 | -0.408 | 0.816 | -0.904 |
| 02 | -0.224 | -0.137 | 0.675 | -1.01 |
| 03 | 0.401 | -0.17 | -0.048 | -0.343 |
| 04 | -0.343 | -2.428 | 1.112 | -1.824 |
| 05 | -0.904 | -2.887 | 1.097 | -0.61 |
| 06 | -0.452 | -3.34 | 1.195 | -3.762 |
| 07 | -3.232 | -3.232 | 1.355 | -3.134 |
| 08 | -3.462 | -3.232 | 1.362 | -3.762 |
| 09 | -1.384 | 0.973 | -3.762 | 0.076 |
| 10 | -2.299 | -2.577 | 1.307 | -2.049 |
| 11 | -3.34 | -1.486 | 0.885 | 0.274 |
| 12 | -1.799 | -3.462 | 1.287 | -1.706 |
| 13 | -1.336 | -2.221 | 1.111 | -0.525 |
| 14 | -0.874 | 1.082 | -0.987 | -1.352 |
| 15 | -0.68 | 0.564 | -0.366 | 0.042 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.56506 |
| 0.0005 | 4.64081 |
| 0.0001 | 6.87506 |