| Motif | JUND.H14INVIVO.0.PM.A |
| Gene (human) | JUND (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Jund |
| Gene synonyms (mouse) | Jun-d, Jund1 |
| LOGO | 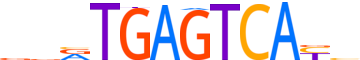 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif | JUND.H14INVIVO.0.PM.A |
| Gene (human) | JUND (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Jund |
| Gene synonyms (mouse) | Jun-d, Jund1 |
| LOGO | 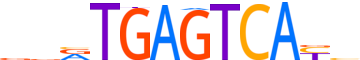 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 12 |
| Consensus | vvRTGAGTCAYh |
| GC content | 48.69% |
| Information content (bits; total / per base) | 14.335 / 1.195 |
| Data sources | ChIP-Seq + Methyl-HT-SELEX |
| Aligned words | 1000 |
| Previous names | JUND.H12INVIVO.0.PM.A |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 25 (163) | 0.87 | 0.987 | 0.839 | 0.972 | 0.873 | 0.988 | 4.004 | 5.759 | 285.319 | 666.456 |
| Mouse | 28 (176) | 0.91 | 0.983 | 0.869 | 0.97 | 0.902 | 0.984 | 3.976 | 5.688 | 281.957 | 648.174 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.838 | 0.811 | 0.708 | 0.697 | 0.622 | 0.625 |
| best | 0.842 | 0.811 | 0.717 | 0.701 | 0.628 | 0.629 | |
| Methyl HT-SELEX, 1 experiments | median | 0.834 | 0.811 | 0.699 | 0.692 | 0.616 | 0.621 |
| best | 0.834 | 0.811 | 0.699 | 0.692 | 0.616 | 0.621 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.842 | 0.811 | 0.717 | 0.701 | 0.628 | 0.629 |
| best | 0.842 | 0.811 | 0.717 | 0.701 | 0.628 | 0.629 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 91.989 | 65.684 | 0.277 | 0.412 |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.486 | 0.059 | 0.369 | 0.091 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic leucine zipper factors (bZIP) {1.1} (TFClass) |
| TF family | Jun-related {1.1.1} (TFClass) |
| TF subfamily | Jun {1.1.1.1} (TFClass) |
| TFClass ID | TFClass: 1.1.1.1.3 |
| HGNC | HGNC:6206 |
| MGI | MGI:96648 |
| EntrezGene (human) | GeneID:3727 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:16478 (SSTAR profile) |
| UniProt ID (human) | JUND_HUMAN |
| UniProt ID (mouse) | JUND_MOUSE |
| UniProt AC (human) | P17535 (TFClass) |
| UniProt AC (mouse) | P15066 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 25 human, 28 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | JUND.H14INVIVO.0.PM.A.pcm |
| PWM | JUND.H14INVIVO.0.PM.A.pwm |
| PFM | JUND.H14INVIVO.0.PM.A.pfm |
| Threshold to P-value map | JUND.H14INVIVO.0.PM.A.thr |
| Motif in other formats | |
| JASPAR format | JUND.H14INVIVO.0.PM.A_jaspar_format.txt |
| MEME format | JUND.H14INVIVO.0.PM.A_meme_format.meme |
| Transfac format | JUND.H14INVIVO.0.PM.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 173.0 | 202.0 | 464.0 | 161.0 |
| 02 | 292.0 | 170.0 | 379.0 | 159.0 |
| 03 | 441.0 | 180.0 | 370.0 | 9.0 |
| 04 | 1.0 | 4.0 | 3.0 | 992.0 |
| 05 | 3.0 | 3.0 | 970.0 | 24.0 |
| 06 | 983.0 | 2.0 | 2.0 | 13.0 |
| 07 | 45.0 | 0.0 | 955.0 | 0.0 |
| 08 | 4.0 | 6.0 | 3.0 | 987.0 |
| 09 | 9.0 | 987.0 | 2.0 | 2.0 |
| 10 | 994.0 | 0.0 | 2.0 | 4.0 |
| 11 | 5.0 | 422.0 | 143.0 | 430.0 |
| 12 | 182.0 | 430.0 | 144.0 | 244.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.173 | 0.202 | 0.464 | 0.161 |
| 02 | 0.292 | 0.17 | 0.379 | 0.159 |
| 03 | 0.441 | 0.18 | 0.37 | 0.009 |
| 04 | 0.001 | 0.004 | 0.003 | 0.992 |
| 05 | 0.003 | 0.003 | 0.97 | 0.024 |
| 06 | 0.983 | 0.002 | 0.002 | 0.013 |
| 07 | 0.045 | 0.0 | 0.955 | 0.0 |
| 08 | 0.004 | 0.006 | 0.003 | 0.987 |
| 09 | 0.009 | 0.987 | 0.002 | 0.002 |
| 10 | 0.994 | 0.0 | 0.002 | 0.004 |
| 11 | 0.005 | 0.422 | 0.143 | 0.43 |
| 12 | 0.182 | 0.43 | 0.144 | 0.244 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.365 | -0.212 | 0.615 | -0.436 |
| 02 | 0.154 | -0.382 | 0.414 | -0.449 |
| 03 | 0.565 | -0.326 | 0.39 | -3.156 |
| 04 | -4.525 | -3.783 | -3.975 | 1.373 |
| 05 | -3.975 | -3.975 | 1.351 | -2.281 |
| 06 | 1.364 | -4.213 | -4.213 | -2.839 |
| 07 | -1.684 | -4.982 | 1.335 | -4.982 |
| 08 | -3.783 | -3.484 | -3.975 | 1.368 |
| 09 | -3.156 | 1.368 | -4.213 | -4.213 |
| 10 | 1.375 | -4.982 | -4.213 | -3.783 |
| 11 | -3.622 | 0.521 | -0.553 | 0.539 |
| 12 | -0.315 | 0.539 | -0.547 | -0.024 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.266065 |
| 0.0005 | 4.05811 |
| 0.0001 | 6.26208 |