| Motif | HMX3.H14INVITRO.0.S.B |
| Gene (human) | HMX3 (GeneCards) |
| Gene synonyms (human) | NKX-5.1, NKX5-1 |
| Gene (mouse) | Hmx3 |
| Gene synonyms (mouse) | Nkx-5.1, Nkx5-1 |
| LOGO | 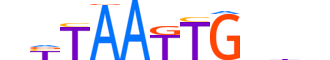 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | HMX3.H14INVITRO.0.S.B |
| Gene (human) | HMX3 (GeneCards) |
| Gene synonyms (human) | NKX-5.1, NKX5-1 |
| Gene (mouse) | Hmx3 |
| Gene synonyms (mouse) | Nkx-5.1, Nkx5-1 |
| LOGO | 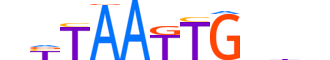 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 11 |
| Consensus | nYTAATTGnbn |
| GC content | 34.42% |
| Information content (bits; total / per base) | 10.054 / 0.914 |
| Data sources | HT-SELEX |
| Aligned words | 9585 |
| Previous names | HMX3.H12INVITRO.0.S.B |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.951 | 0.926 | 0.884 | 0.858 | 0.745 | 0.748 |
| best | 0.975 | 0.96 | 0.93 | 0.911 | 0.785 | 0.791 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.994 | 0.646 | 0.92 | 0.45 |
| batch 2 | 0.863 | 0.186 | 0.779 | 0.462 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Homeo domain factors {3.1} (TFClass) |
| TF family | NK-related {3.1.2} (TFClass) |
| TF subfamily | NK5-HMX {3.1.2.18} (TFClass) |
| TFClass ID | TFClass: 3.1.2.18.3 |
| HGNC | HGNC:5019 |
| MGI | MGI:107160 |
| EntrezGene (human) | GeneID:340784 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:15373 (SSTAR profile) |
| UniProt ID (human) | HMX3_HUMAN |
| UniProt ID (mouse) | HMX3_MOUSE |
| UniProt AC (human) | A6NHT5 (TFClass) |
| UniProt AC (mouse) | P42581 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | HMX3.H14INVITRO.0.S.B.pcm |
| PWM | HMX3.H14INVITRO.0.S.B.pwm |
| PFM | HMX3.H14INVITRO.0.S.B.pfm |
| Threshold to P-value map | HMX3.H14INVITRO.0.S.B.thr |
| Motif in other formats | |
| JASPAR format | HMX3.H14INVITRO.0.S.B_jaspar_format.txt |
| MEME format | HMX3.H14INVITRO.0.S.B_meme_format.meme |
| Transfac format | HMX3.H14INVITRO.0.S.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 2352.0 | 2356.0 | 2865.0 | 2012.0 |
| 02 | 1008.5 | 1425.5 | 1054.5 | 6096.5 |
| 03 | 172.0 | 1047.0 | 118.0 | 8248.0 |
| 04 | 9391.0 | 2.0 | 192.0 | 0.0 |
| 05 | 9585.0 | 0.0 | 0.0 | 0.0 |
| 06 | 3.0 | 0.0 | 2602.0 | 6980.0 |
| 07 | 6.0 | 1166.0 | 14.0 | 8399.0 |
| 08 | 571.0 | 34.0 | 8970.0 | 10.0 |
| 09 | 2042.0 | 3650.0 | 2108.0 | 1785.0 |
| 10 | 1484.25 | 2647.25 | 1557.25 | 3896.25 |
| 11 | 2300.0 | 2643.0 | 1841.0 | 2801.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.245 | 0.246 | 0.299 | 0.21 |
| 02 | 0.105 | 0.149 | 0.11 | 0.636 |
| 03 | 0.018 | 0.109 | 0.012 | 0.861 |
| 04 | 0.98 | 0.0 | 0.02 | 0.0 |
| 05 | 1.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.271 | 0.728 |
| 07 | 0.001 | 0.122 | 0.001 | 0.876 |
| 08 | 0.06 | 0.004 | 0.936 | 0.001 |
| 09 | 0.213 | 0.381 | 0.22 | 0.186 |
| 10 | 0.155 | 0.276 | 0.162 | 0.406 |
| 11 | 0.24 | 0.276 | 0.192 | 0.292 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.019 | -0.017 | 0.179 | -0.175 |
| 02 | -0.864 | -0.519 | -0.82 | 0.933 |
| 03 | -2.622 | -0.827 | -2.993 | 1.235 |
| 04 | 1.365 | -6.326 | -2.513 | -6.953 |
| 05 | 1.386 | -6.953 | -6.953 | -6.953 |
| 06 | -6.116 | -6.953 | 0.082 | 1.069 |
| 07 | -5.667 | -0.719 | -4.992 | 1.254 |
| 08 | -1.431 | -4.191 | 1.319 | -5.274 |
| 09 | -0.16 | 0.42 | -0.128 | -0.294 |
| 10 | -0.478 | 0.1 | -0.43 | 0.486 |
| 11 | -0.041 | 0.098 | -0.263 | 0.156 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.89167 |
| 0.0005 | 5.9438 |
| 0.0001 | 7.573335 |