| Motif | GLIS1.H14INVIVO.0.P.B |
| Gene (human) | GLIS1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Glis1 |
| Gene synonyms (mouse) | Gli5 |
| LOGO |  |
| LOGO (reverse complement) | 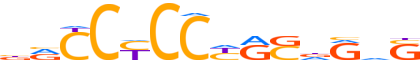 |
| Motif subtype | 0 |
| Quality | B |
| Motif | GLIS1.H14INVIVO.0.P.B |
| Gene (human) | GLIS1 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | Glis1 |
| Gene synonyms (mouse) | Gli5 |
| LOGO |  |
| LOGO (reverse complement) | 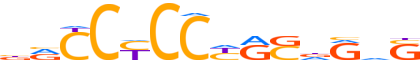 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 14 |
| Consensus | SdYhSYRGGRGGYv |
| GC content | 71.97% |
| Information content (bits; total / per base) | 11.907 / 0.851 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| Previous names | GLIS1.H12INVIVO.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 2 (12) | 0.868 | 0.92 | 0.743 | 0.838 | 0.795 | 0.833 | 2.381 | 2.577 | 130.436 | 279.456 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 2 experiments | median | 0.956 | 0.94 | 0.876 | 0.866 | 0.753 | 0.766 |
| best | 0.992 | 0.988 | 0.975 | 0.966 | 0.873 | 0.872 | |
| Methyl HT-SELEX, 1 experiments | median | 0.992 | 0.988 | 0.975 | 0.966 | 0.873 | 0.872 |
| best | 0.992 | 0.988 | 0.975 | 0.966 | 0.873 | 0.872 | |
| Non-Methyl HT-SELEX, 1 experiments | median | 0.919 | 0.892 | 0.777 | 0.767 | 0.633 | 0.66 |
| best | 0.919 | 0.892 | 0.777 | 0.767 | 0.633 | 0.66 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.711 | 0.388 | 0.586 | 0.491 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | C2H2 zinc finger factors {2.3} (TFClass) |
| TF family | More than 3 adjacent zinc fingers {2.3.3} (TFClass) |
| TF subfamily | GLI-like {2.3.3.1} (TFClass) |
| TFClass ID | TFClass: 2.3.3.1.4 |
| HGNC | HGNC:29525 |
| MGI | MGI:2386723 |
| EntrezGene (human) | GeneID:148979 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:230587 (SSTAR profile) |
| UniProt ID (human) | GLIS1_HUMAN |
| UniProt ID (mouse) | GLIS1_MOUSE |
| UniProt AC (human) | Q8NBF1 (TFClass) |
| UniProt AC (mouse) | Q8K1M4 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 2 human, 0 mouse |
| HT-SELEX | 1 |
| Methyl-HT-SELEX | 1 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | GLIS1.H14INVIVO.0.P.B.pcm |
| PWM | GLIS1.H14INVIVO.0.P.B.pwm |
| PFM | GLIS1.H14INVIVO.0.P.B.pfm |
| Threshold to P-value map | GLIS1.H14INVIVO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | GLIS1.H14INVIVO.0.P.B_jaspar_format.txt |
| MEME format | GLIS1.H14INVIVO.0.P.B_meme_format.meme |
| Transfac format | GLIS1.H14INVIVO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 97.0 | 693.0 | 196.0 | 14.0 |
| 02 | 213.0 | 143.0 | 217.0 | 427.0 |
| 03 | 40.0 | 729.0 | 111.0 | 120.0 |
| 04 | 175.0 | 335.0 | 59.0 | 431.0 |
| 05 | 8.0 | 414.0 | 558.0 | 20.0 |
| 06 | 15.0 | 593.0 | 16.0 | 376.0 |
| 07 | 329.0 | 4.0 | 493.0 | 174.0 |
| 08 | 8.0 | 2.0 | 925.0 | 65.0 |
| 09 | 1.0 | 1.0 | 992.0 | 6.0 |
| 10 | 475.0 | 4.0 | 457.0 | 64.0 |
| 11 | 9.0 | 2.0 | 987.0 | 2.0 |
| 12 | 114.0 | 14.0 | 825.0 | 47.0 |
| 13 | 14.0 | 479.0 | 162.0 | 345.0 |
| 14 | 205.0 | 483.0 | 182.0 | 130.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.097 | 0.693 | 0.196 | 0.014 |
| 02 | 0.213 | 0.143 | 0.217 | 0.427 |
| 03 | 0.04 | 0.729 | 0.111 | 0.12 |
| 04 | 0.175 | 0.335 | 0.059 | 0.431 |
| 05 | 0.008 | 0.414 | 0.558 | 0.02 |
| 06 | 0.015 | 0.593 | 0.016 | 0.376 |
| 07 | 0.329 | 0.004 | 0.493 | 0.174 |
| 08 | 0.008 | 0.002 | 0.925 | 0.065 |
| 09 | 0.001 | 0.001 | 0.992 | 0.006 |
| 10 | 0.475 | 0.004 | 0.457 | 0.064 |
| 11 | 0.009 | 0.002 | 0.987 | 0.002 |
| 12 | 0.114 | 0.014 | 0.825 | 0.047 |
| 13 | 0.014 | 0.479 | 0.162 | 0.345 |
| 14 | 0.205 | 0.483 | 0.182 | 0.13 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.936 | 1.015 | -0.241 | -2.773 |
| 02 | -0.159 | -0.553 | -0.141 | 0.532 |
| 03 | -1.797 | 1.066 | -0.803 | -0.727 |
| 04 | -0.354 | 0.291 | -1.422 | 0.542 |
| 05 | -3.253 | 0.502 | 0.799 | -2.45 |
| 06 | -2.711 | 0.86 | -2.653 | 0.406 |
| 07 | 0.273 | -3.783 | 0.676 | -0.359 |
| 08 | -3.253 | -4.213 | 1.303 | -1.328 |
| 09 | -4.525 | -4.525 | 1.373 | -3.484 |
| 10 | 0.639 | -3.783 | 0.6 | -1.343 |
| 11 | -3.156 | -4.213 | 1.368 | -4.213 |
| 12 | -0.777 | -2.773 | 1.189 | -1.642 |
| 13 | -2.773 | 0.647 | -0.43 | 0.32 |
| 14 | -0.197 | 0.655 | -0.315 | -0.648 |
| P-value | Threshold |
|---|---|
| 0.001 | 4.15626 |
| 0.0005 | 5.20586 |
| 0.0001 | 7.30916 |