| Motif | FOXL1.H14RSNP.0.S.D |
| Gene (human) | FOXL1 (GeneCards) |
| Gene synonyms (human) | FKHL11, FREAC7 |
| Gene (mouse) | Foxl1 |
| Gene synonyms (mouse) | Fkh6, Fkhl11 |
| LOGO | 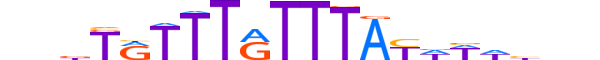 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif | FOXL1.H14RSNP.0.S.D |
| Gene (human) | FOXL1 (GeneCards) |
| Gene synonyms (human) | FKHL11, FREAC7 |
| Gene (mouse) | Foxl1 |
| Gene synonyms (mouse) | Fkh6, Fkhl11 |
| LOGO |  |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 20 |
| Consensus | bndTRTTTGTTTAYWWdhnn |
| GC content | 26.87% |
| Information content (bits; total / per base) | 17.812 / 0.891 |
| Data sources | HT-SELEX |
| Aligned words | 7137 |
| Previous names | FOXL1.H12RSNP.0.S.D |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.993 | 0.99 | 0.911 | 0.917 | 0.711 | 0.764 |
| best | 0.999 | 0.999 | 0.99 | 0.988 | 0.782 | 0.832 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Fork head/winged helix factors {3.3} (TFClass) |
| TF family | FOX {3.3.1} (TFClass) |
| TF subfamily | FOXL {3.3.1.12} (TFClass) |
| TFClass ID | TFClass: 3.3.1.12.1 |
| HGNC | HGNC:3817 |
| MGI | MGI:1347469 |
| EntrezGene (human) | GeneID:2300 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:14241 (SSTAR profile) |
| UniProt ID (human) | FOXL1_HUMAN |
| UniProt ID (mouse) | FOXL1_MOUSE |
| UniProt AC (human) | Q12952 (TFClass) |
| UniProt AC (mouse) | Q64731 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 0 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | FOXL1.H14RSNP.0.S.D.pcm |
| PWM | FOXL1.H14RSNP.0.S.D.pwm |
| PFM | FOXL1.H14RSNP.0.S.D.pfm |
| Threshold to P-value map | FOXL1.H14RSNP.0.S.D.thr |
| Motif in other formats | |
| JASPAR format | FOXL1.H14RSNP.0.S.D_jaspar_format.txt |
| MEME format | FOXL1.H14RSNP.0.S.D_meme_format.meme |
| Transfac format | FOXL1.H14RSNP.0.S.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1225.0 | 1327.0 | 2811.0 | 1774.0 |
| 02 | 1486.75 | 1067.75 | 2359.75 | 2222.75 |
| 03 | 1182.0 | 788.0 | 2048.0 | 3119.0 |
| 04 | 278.0 | 370.0 | 482.0 | 6007.0 |
| 05 | 1359.0 | 422.0 | 4915.0 | 441.0 |
| 06 | 641.0 | 103.0 | 183.0 | 6210.0 |
| 07 | 308.0 | 25.0 | 39.0 | 6765.0 |
| 08 | 0.0 | 1.0 | 1.0 | 7135.0 |
| 09 | 2961.0 | 1.0 | 4175.0 | 0.0 |
| 10 | 0.0 | 19.0 | 0.0 | 7118.0 |
| 11 | 9.0 | 3.0 | 3.0 | 7122.0 |
| 12 | 11.0 | 2.0 | 171.0 | 6953.0 |
| 13 | 6521.0 | 30.0 | 449.0 | 137.0 |
| 14 | 279.0 | 2599.0 | 88.0 | 4171.0 |
| 15 | 2406.0 | 496.0 | 572.0 | 3663.0 |
| 16 | 1377.0 | 555.0 | 500.0 | 4705.0 |
| 17 | 2538.0 | 522.0 | 547.0 | 3530.0 |
| 18 | 1211.0 | 2296.0 | 760.0 | 2870.0 |
| 19 | 1542.5 | 2436.5 | 1204.5 | 1953.5 |
| 20 | 1377.0 | 2704.0 | 1284.0 | 1772.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.172 | 0.186 | 0.394 | 0.249 |
| 02 | 0.208 | 0.15 | 0.331 | 0.311 |
| 03 | 0.166 | 0.11 | 0.287 | 0.437 |
| 04 | 0.039 | 0.052 | 0.068 | 0.842 |
| 05 | 0.19 | 0.059 | 0.689 | 0.062 |
| 06 | 0.09 | 0.014 | 0.026 | 0.87 |
| 07 | 0.043 | 0.004 | 0.005 | 0.948 |
| 08 | 0.0 | 0.0 | 0.0 | 1.0 |
| 09 | 0.415 | 0.0 | 0.585 | 0.0 |
| 10 | 0.0 | 0.003 | 0.0 | 0.997 |
| 11 | 0.001 | 0.0 | 0.0 | 0.998 |
| 12 | 0.002 | 0.0 | 0.024 | 0.974 |
| 13 | 0.914 | 0.004 | 0.063 | 0.019 |
| 14 | 0.039 | 0.364 | 0.012 | 0.584 |
| 15 | 0.337 | 0.069 | 0.08 | 0.513 |
| 16 | 0.193 | 0.078 | 0.07 | 0.659 |
| 17 | 0.356 | 0.073 | 0.077 | 0.495 |
| 18 | 0.17 | 0.322 | 0.106 | 0.402 |
| 19 | 0.216 | 0.341 | 0.169 | 0.274 |
| 20 | 0.193 | 0.379 | 0.18 | 0.248 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.375 | -0.296 | 0.454 | -0.006 |
| 02 | -0.182 | -0.513 | 0.279 | 0.22 |
| 03 | -0.411 | -0.816 | 0.138 | 0.558 |
| 04 | -1.852 | -1.569 | -1.305 | 1.213 |
| 05 | -0.272 | -1.438 | 1.013 | -1.394 |
| 06 | -1.022 | -2.832 | -2.266 | 1.246 |
| 07 | -1.751 | -4.184 | -3.769 | 1.332 |
| 08 | -6.691 | -6.319 | -6.319 | 1.385 |
| 09 | 0.506 | -6.319 | 0.849 | -6.691 |
| 10 | -6.691 | -4.433 | -6.691 | 1.383 |
| 11 | -5.07 | -5.836 | -5.836 | 1.383 |
| 12 | -4.906 | -6.049 | -2.333 | 1.359 |
| 13 | 1.295 | -4.015 | -1.376 | -2.552 |
| 14 | -1.849 | 0.376 | -2.986 | 0.848 |
| 15 | 0.299 | -1.277 | -1.135 | 0.719 |
| 16 | -0.259 | -1.165 | -1.269 | 0.969 |
| 17 | 0.352 | -1.226 | -1.179 | 0.682 |
| 18 | -0.387 | 0.252 | -0.852 | 0.475 |
| 19 | -0.145 | 0.311 | -0.392 | 0.091 |
| 20 | -0.259 | 0.415 | -0.329 | -0.007 |
| P-value | Threshold |
|---|---|
| 0.001 | 0.19491 |
| 0.0005 | 1.83511 |
| 0.0001 | 5.20296 |