| Motif | FOXD3.H14RSNP.0.P.D |
| Gene (human) | FOXD3 (GeneCards) |
| Gene synonyms (human) | HFH2 |
| Gene (mouse) | Foxd3 |
| Gene synonyms (mouse) | Hfh2 |
| LOGO |  |
| LOGO (reverse complement) | 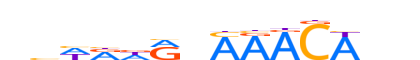 |
| Motif subtype | 0 |
| Quality | D |
| Motif | FOXD3.H14RSNP.0.P.D |
| Gene (human) | FOXD3 (GeneCards) |
| Gene synonyms (human) | HFH2 |
| Gene (mouse) | Foxd3 |
| Gene synonyms (mouse) | Hfh2 |
| LOGO |  |
| LOGO (reverse complement) | 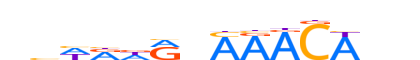 |
| Motif subtype | 0 |
| Quality | D |
| Motif length | 14 |
| Consensus | nnTGTYKnYhYWvn |
| GC content | 39.12% |
| Information content (bits; total / per base) | 7.704 / 0.55 |
| Data sources | ChIP-Seq |
| Aligned words | 997 |
| Previous names | FOXD3.H12RSNP.0.P.D |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 1 (7) | 0.684 | 0.717 | 0.495 | 0.559 | 0.753 | 0.786 | 2.175 | 2.496 | 62.921 | 89.347 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.626 | 0.614 | 0.57 | 0.566 | 0.535 | 0.54 |
| best | 0.71 | 0.693 | 0.617 | 0.613 | 0.558 | 0.567 | |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Fork head/winged helix factors {3.3} (TFClass) |
| TF family | FOX {3.3.1} (TFClass) |
| TF subfamily | FOXD {3.3.1.4} (TFClass) |
| TFClass ID | TFClass: 3.3.1.4.3 |
| HGNC | HGNC:3804 |
| MGI | MGI:1347473 |
| EntrezGene (human) | GeneID:27022 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:15221 (SSTAR profile) |
| UniProt ID (human) | FOXD3_HUMAN |
| UniProt ID (mouse) | FOXD3_MOUSE |
| UniProt AC (human) | Q9UJU5 (TFClass) |
| UniProt AC (mouse) | Q61060 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 1 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | FOXD3.H14RSNP.0.P.D.pcm |
| PWM | FOXD3.H14RSNP.0.P.D.pwm |
| PFM | FOXD3.H14RSNP.0.P.D.pfm |
| Threshold to P-value map | FOXD3.H14RSNP.0.P.D.thr |
| Motif in other formats | |
| JASPAR format | FOXD3.H14RSNP.0.P.D_jaspar_format.txt |
| MEME format | FOXD3.H14RSNP.0.P.D_meme_format.meme |
| Transfac format | FOXD3.H14RSNP.0.P.D_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 196.0 | 196.0 | 303.0 | 302.0 |
| 02 | 152.0 | 269.0 | 242.0 | 334.0 |
| 03 | 177.0 | 10.0 | 44.0 | 766.0 |
| 04 | 36.0 | 75.0 | 880.0 | 6.0 |
| 05 | 61.0 | 72.0 | 29.0 | 835.0 |
| 06 | 43.0 | 89.0 | 63.0 | 802.0 |
| 07 | 50.0 | 7.0 | 180.0 | 760.0 |
| 08 | 324.0 | 167.0 | 253.0 | 253.0 |
| 09 | 16.0 | 600.0 | 53.0 | 328.0 |
| 10 | 274.0 | 196.0 | 40.0 | 487.0 |
| 11 | 93.0 | 234.0 | 80.0 | 590.0 |
| 12 | 438.0 | 29.0 | 115.0 | 415.0 |
| 13 | 233.0 | 165.0 | 436.0 | 163.0 |
| 14 | 135.0 | 308.0 | 326.0 | 228.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.197 | 0.197 | 0.304 | 0.303 |
| 02 | 0.152 | 0.27 | 0.243 | 0.335 |
| 03 | 0.178 | 0.01 | 0.044 | 0.768 |
| 04 | 0.036 | 0.075 | 0.883 | 0.006 |
| 05 | 0.061 | 0.072 | 0.029 | 0.838 |
| 06 | 0.043 | 0.089 | 0.063 | 0.804 |
| 07 | 0.05 | 0.007 | 0.181 | 0.762 |
| 08 | 0.325 | 0.168 | 0.254 | 0.254 |
| 09 | 0.016 | 0.602 | 0.053 | 0.329 |
| 10 | 0.275 | 0.197 | 0.04 | 0.488 |
| 11 | 0.093 | 0.235 | 0.08 | 0.592 |
| 12 | 0.439 | 0.029 | 0.115 | 0.416 |
| 13 | 0.234 | 0.165 | 0.437 | 0.163 |
| 14 | 0.135 | 0.309 | 0.327 | 0.229 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.238 | -0.238 | 0.194 | 0.191 |
| 02 | -0.49 | 0.076 | -0.029 | 0.291 |
| 03 | -0.34 | -3.064 | -1.703 | 1.118 |
| 04 | -1.895 | -1.185 | 1.257 | -3.481 |
| 05 | -1.387 | -1.225 | -2.1 | 1.204 |
| 06 | -1.725 | -1.018 | -1.355 | 1.164 |
| 07 | -1.579 | -3.359 | -0.323 | 1.11 |
| 08 | 0.261 | -0.397 | 0.015 | 0.015 |
| 09 | -2.65 | 0.874 | -1.523 | 0.273 |
| 10 | 0.094 | -0.238 | -1.794 | 0.666 |
| 11 | -0.974 | -0.063 | -1.122 | 0.858 |
| 12 | 0.561 | -2.1 | -0.766 | 0.507 |
| 13 | -0.067 | -0.409 | 0.556 | -0.421 |
| 14 | -0.607 | 0.21 | 0.267 | -0.088 |
| P-value | Threshold |
|---|---|
| 0.001 | 5.00091 |
| 0.0005 | 5.66906 |
| 0.0001 | 7.02146 |