| Motif | FOSL2.H14CORE.0.P.B |
| Gene (human) | FOSL2 (GeneCards) |
| Gene synonyms (human) | FRA2 |
| Gene (mouse) | Fosl2 |
| Gene synonyms (mouse) | Fra-2, Fra2 |
| LOGO | 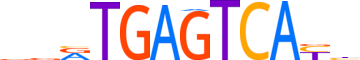 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif | FOSL2.H14CORE.0.P.B |
| Gene (human) | FOSL2 (GeneCards) |
| Gene synonyms (human) | FRA2 |
| Gene (mouse) | Fosl2 |
| Gene synonyms (mouse) | Fra-2, Fra2 |
| LOGO | 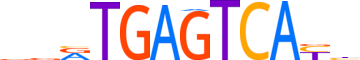 |
| LOGO (reverse complement) |  |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 12 |
| Consensus | vvRTGAGTCAYh |
| GC content | 48.13% |
| Information content (bits; total / per base) | 14.649 / 1.221 |
| Data sources | ChIP-Seq |
| Aligned words | 1001 |
| Previous names | FOSL2.H12CORE.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 28 (191) | 0.963 | 0.992 | 0.918 | 0.978 | 0.964 | 0.987 | 4.437 | 5.341 | 329.569 | 750.77 |
| Mouse | 11 (71) | 0.96 | 0.992 | 0.921 | 0.978 | 0.959 | 0.991 | 4.381 | 5.391 | 441.495 | 656.886 |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 100.0 | 4.076 | 0.357 | 0.272 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic leucine zipper factors (bZIP) {1.1} (TFClass) |
| TF family | Fos-related {1.1.2} (TFClass) |
| TF subfamily | Fos {1.1.2.1} (TFClass) |
| TFClass ID | TFClass: 1.1.2.1.4 |
| HGNC | HGNC:3798 |
| MGI | MGI:102858 |
| EntrezGene (human) | GeneID:2355 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:14284 (SSTAR profile) |
| UniProt ID (human) | FOSL2_HUMAN |
| UniProt ID (mouse) | FOSL2_MOUSE |
| UniProt AC (human) | P15408 (TFClass) |
| UniProt AC (mouse) | P47930 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 28 human, 11 mouse |
| HT-SELEX | 0 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | FOSL2.H14CORE.0.P.B.pcm |
| PWM | FOSL2.H14CORE.0.P.B.pwm |
| PFM | FOSL2.H14CORE.0.P.B.pfm |
| Threshold to P-value map | FOSL2.H14CORE.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | FOSL2.H14CORE.0.P.B_jaspar_format.txt |
| MEME format | FOSL2.H14CORE.0.P.B_meme_format.meme |
| Transfac format | FOSL2.H14CORE.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 190.0 | 211.0 | 419.0 | 181.0 |
| 02 | 232.0 | 176.0 | 433.0 | 160.0 |
| 03 | 488.0 | 152.0 | 346.0 | 15.0 |
| 04 | 0.0 | 3.0 | 2.0 | 996.0 |
| 05 | 2.0 | 1.0 | 982.0 | 16.0 |
| 06 | 986.0 | 6.0 | 0.0 | 9.0 |
| 07 | 48.0 | 0.0 | 953.0 | 0.0 |
| 08 | 0.0 | 1.0 | 0.0 | 1000.0 |
| 09 | 5.0 | 995.0 | 1.0 | 0.0 |
| 10 | 998.0 | 1.0 | 0.0 | 2.0 |
| 11 | 10.0 | 372.0 | 131.0 | 488.0 |
| 12 | 164.0 | 451.0 | 145.0 | 241.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.19 | 0.211 | 0.419 | 0.181 |
| 02 | 0.232 | 0.176 | 0.433 | 0.16 |
| 03 | 0.488 | 0.152 | 0.346 | 0.015 |
| 04 | 0.0 | 0.003 | 0.002 | 0.995 |
| 05 | 0.002 | 0.001 | 0.981 | 0.016 |
| 06 | 0.985 | 0.006 | 0.0 | 0.009 |
| 07 | 0.048 | 0.0 | 0.952 | 0.0 |
| 08 | 0.0 | 0.001 | 0.0 | 0.999 |
| 09 | 0.005 | 0.994 | 0.001 | 0.0 |
| 10 | 0.997 | 0.001 | 0.0 | 0.002 |
| 11 | 0.01 | 0.372 | 0.131 | 0.488 |
| 12 | 0.164 | 0.451 | 0.145 | 0.241 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.273 | -0.169 | 0.513 | -0.321 |
| 02 | -0.075 | -0.349 | 0.545 | -0.443 |
| 03 | 0.665 | -0.494 | 0.322 | -2.712 |
| 04 | -4.983 | -3.976 | -4.214 | 1.376 |
| 05 | -4.214 | -4.526 | 1.362 | -2.654 |
| 06 | 1.366 | -3.485 | -4.983 | -3.157 |
| 07 | -1.623 | -4.983 | 1.332 | -4.983 |
| 08 | -4.983 | -4.526 | -4.983 | 1.38 |
| 09 | -3.623 | 1.375 | -4.526 | -4.983 |
| 10 | 1.378 | -4.526 | -4.983 | -4.214 |
| 11 | -3.067 | 0.394 | -0.641 | 0.665 |
| 12 | -0.419 | 0.586 | -0.541 | -0.037 |
| P-value | Threshold |
|---|---|
| 0.001 | 1.95841 |
| 0.0005 | 3.66962 |
| 0.0001 | 6.12323 |