| Motif | E2F4.H14RSNP.0.P.B |
| Gene (human) | E2F4 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | E2f4 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 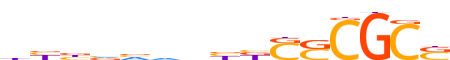 |
| Motif subtype | 0 |
| Quality | B |
| Motif | E2F4.H14RSNP.0.P.B |
| Gene (human) | E2F4 (GeneCards) |
| Gene synonyms (human) | |
| Gene (mouse) | E2f4 |
| Gene synonyms (mouse) | |
| LOGO |  |
| LOGO (reverse complement) | 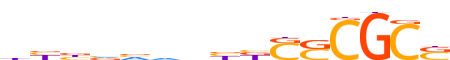 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 15 |
| Consensus | SGCGSSvvndbbvvv |
| GC content | 67.24% |
| Information content (bits; total / per base) | 8.309 / 0.554 |
| Data sources | ChIP-Seq |
| Aligned words | 1018 |
| Previous names | E2F4.H12RSNP.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 8 (56) | 0.817 | 0.873 | 0.666 | 0.791 | 0.78 | 0.828 | 2.394 | 3.224 | 136.342 | 250.432 |
| Mouse | 12 (81) | 0.813 | 0.853 | 0.64 | 0.696 | 0.723 | 0.822 | 2.074 | 2.554 | 106.167 | 250.42 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 2 experiments | median | 0.717 | 0.694 | 0.711 | 0.687 | 0.697 | 0.674 |
| best | 0.935 | 0.888 | 0.925 | 0.876 | 0.897 | 0.849 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 1 | 0.645 | 0.098 | 0.403 | 0.288 |
| TF superclass | Helix-turn-helix domains {3} (TFClass) |
| TF class | Fork head/winged helix factors {3.3} (TFClass) |
| TF family | E2F {3.3.2} (TFClass) |
| TF subfamily | E2F {3.3.2.1} (TFClass) |
| TFClass ID | TFClass: 3.3.2.1.4 |
| HGNC | HGNC:3118 |
| MGI | MGI:103012 |
| EntrezGene (human) | GeneID:1874 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:104394 (SSTAR profile) |
| UniProt ID (human) | E2F4_HUMAN |
| UniProt ID (mouse) | E2F4_MOUSE |
| UniProt AC (human) | Q16254 (TFClass) |
| UniProt AC (mouse) | Q8R0K9 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 8 human, 12 mouse |
| HT-SELEX | 2 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | E2F4.H14RSNP.0.P.B.pcm |
| PWM | E2F4.H14RSNP.0.P.B.pwm |
| PFM | E2F4.H14RSNP.0.P.B.pfm |
| Threshold to P-value map | E2F4.H14RSNP.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | E2F4.H14RSNP.0.P.B_jaspar_format.txt |
| MEME format | E2F4.H14RSNP.0.P.B_meme_format.meme |
| Transfac format | E2F4.H14RSNP.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 84.0 | 341.0 | 551.0 | 42.0 |
| 02 | 9.0 | 98.0 | 894.0 | 17.0 |
| 03 | 30.0 | 967.0 | 5.0 | 16.0 |
| 04 | 25.0 | 47.0 | 931.0 | 15.0 |
| 05 | 40.0 | 426.0 | 536.0 | 16.0 |
| 06 | 53.0 | 206.0 | 726.0 | 33.0 |
| 07 | 504.0 | 264.0 | 212.0 | 38.0 |
| 08 | 460.0 | 130.0 | 335.0 | 93.0 |
| 09 | 316.0 | 286.0 | 218.0 | 198.0 |
| 10 | 179.0 | 152.0 | 251.0 | 436.0 |
| 11 | 69.0 | 217.0 | 282.0 | 450.0 |
| 12 | 72.0 | 346.0 | 292.0 | 308.0 |
| 13 | 276.0 | 273.0 | 410.0 | 59.0 |
| 14 | 521.0 | 108.0 | 306.0 | 83.0 |
| 15 | 428.0 | 201.0 | 256.0 | 133.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.083 | 0.335 | 0.541 | 0.041 |
| 02 | 0.009 | 0.096 | 0.878 | 0.017 |
| 03 | 0.029 | 0.95 | 0.005 | 0.016 |
| 04 | 0.025 | 0.046 | 0.915 | 0.015 |
| 05 | 0.039 | 0.418 | 0.527 | 0.016 |
| 06 | 0.052 | 0.202 | 0.713 | 0.032 |
| 07 | 0.495 | 0.259 | 0.208 | 0.037 |
| 08 | 0.452 | 0.128 | 0.329 | 0.091 |
| 09 | 0.31 | 0.281 | 0.214 | 0.194 |
| 10 | 0.176 | 0.149 | 0.247 | 0.428 |
| 11 | 0.068 | 0.213 | 0.277 | 0.442 |
| 12 | 0.071 | 0.34 | 0.287 | 0.303 |
| 13 | 0.271 | 0.268 | 0.403 | 0.058 |
| 14 | 0.512 | 0.106 | 0.301 | 0.082 |
| 15 | 0.42 | 0.197 | 0.251 | 0.131 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -1.095 | 0.291 | 0.769 | -1.768 |
| 02 | -3.173 | -0.944 | 1.252 | -2.616 |
| 03 | -2.089 | 1.33 | -3.639 | -2.671 |
| 04 | -2.26 | -1.66 | 1.292 | -2.729 |
| 05 | -1.815 | 0.512 | 0.741 | -2.671 |
| 06 | -1.544 | -0.21 | 1.044 | -1.998 |
| 07 | 0.68 | 0.036 | -0.181 | -1.864 |
| 08 | 0.589 | -0.665 | 0.273 | -0.995 |
| 09 | 0.215 | 0.116 | -0.154 | -0.249 |
| 10 | -0.349 | -0.511 | -0.014 | 0.536 |
| 11 | -1.287 | -0.158 | 0.102 | 0.567 |
| 12 | -1.246 | 0.305 | 0.137 | 0.19 |
| 13 | 0.081 | 0.07 | 0.474 | -1.44 |
| 14 | 0.713 | -0.848 | 0.183 | -1.107 |
| 15 | 0.517 | -0.234 | 0.006 | -0.643 |
| P-value | Threshold |
|---|---|
| 0.001 | 5.01716 |
| 0.0005 | 5.74346 |
| 0.0001 | 7.17416 |