| Motif | ASCL2.H14RSNP.0.PSM.A |
| Gene (human) | ASCL2 (GeneCards) |
| Gene synonyms (human) | BHLHA45, HASH2 |
| Gene (mouse) | Ascl2 |
| Gene synonyms (mouse) | Mash2 |
| LOGO |  |
| LOGO (reverse complement) | 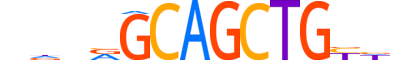 |
| Motif subtype | 0 |
| Quality | A |
| Motif | ASCL2.H14RSNP.0.PSM.A |
| Gene (human) | ASCL2 (GeneCards) |
| Gene synonyms (human) | BHLHA45, HASH2 |
| Gene (mouse) | Ascl2 |
| Gene synonyms (mouse) | Mash2 |
| LOGO |  |
| LOGO (reverse complement) | 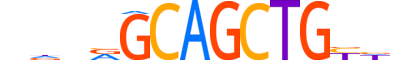 |
| Motif subtype | 0 |
| Quality | A |
| Motif length | 14 |
| Consensus | nhRCAGCTGCYnhn |
| GC content | 58.52% |
| Information content (bits; total / per base) | 14.908 / 1.065 |
| Data sources | ChIP-Seq + HT-SELEX + Methyl-HT-SELEX |
| Aligned words | 359 |
| Previous names | ASCL2.H12RSNP.0.PSM.A |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | 2 (12) | 0.858 | 0.909 | 0.777 | 0.834 | 0.752 | 0.815 | 2.321 | 2.657 | 150.652 | 290.009 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Overall, 6 experiments | median | 0.864 | 0.847 | 0.716 | 0.714 | 0.616 | 0.631 |
| best | 0.963 | 0.947 | 0.868 | 0.85 | 0.731 | 0.736 | |
| Methyl HT-SELEX, 2 experiments | median | 0.894 | 0.876 | 0.75 | 0.744 | 0.637 | 0.651 |
| best | 0.902 | 0.881 | 0.766 | 0.755 | 0.649 | 0.66 | |
| Non-Methyl HT-SELEX, 4 experiments | median | 0.748 | 0.742 | 0.636 | 0.641 | 0.574 | 0.585 |
| best | 0.963 | 0.947 | 0.868 | 0.85 | 0.731 | 0.736 | |
| rSNP benchmarking, SNP-SELEX | auROC | auPRC | Pearson r | Kendall tau |
|---|---|---|---|---|
| batch 2 | 0.706 | 0.429 | 0.717 | 0.512 |
| TF superclass | Basic domains {1} (TFClass) |
| TF class | Basic helix-loop-helix factors (bHLH) {1.2} (TFClass) |
| TF family | MyoD-ASC-related {1.2.2} (TFClass) |
| TF subfamily | ASC {1.2.2.2} (TFClass) |
| TFClass ID | TFClass: 1.2.2.2.2 |
| HGNC | HGNC:739 |
| MGI | MGI:96920 |
| EntrezGene (human) | GeneID:430 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:17173 (SSTAR profile) |
| UniProt ID (human) | ASCL2_HUMAN |
| UniProt ID (mouse) | ASCL2_MOUSE |
| UniProt AC (human) | Q99929 (TFClass) |
| UniProt AC (mouse) | O35885 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 0 human, 2 mouse |
| HT-SELEX | 4 |
| Methyl-HT-SELEX | 2 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | ASCL2.H14RSNP.0.PSM.A.pcm |
| PWM | ASCL2.H14RSNP.0.PSM.A.pwm |
| PFM | ASCL2.H14RSNP.0.PSM.A.pfm |
| Threshold to P-value map | ASCL2.H14RSNP.0.PSM.A.thr |
| Motif in other formats | |
| JASPAR format | ASCL2.H14RSNP.0.PSM.A_jaspar_format.txt |
| MEME format | ASCL2.H14RSNP.0.PSM.A_meme_format.meme |
| Transfac format | ASCL2.H14RSNP.0.PSM.A_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 47.5 | 103.5 | 115.5 | 92.5 |
| 02 | 196.75 | 54.75 | 49.75 | 57.75 |
| 03 | 175.0 | 32.0 | 137.0 | 15.0 |
| 04 | 1.0 | 358.0 | 0.0 | 0.0 |
| 05 | 359.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 4.0 | 355.0 | 0.0 |
| 07 | 1.0 | 358.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 359.0 |
| 09 | 2.0 | 2.0 | 355.0 | 0.0 |
| 10 | 2.0 | 337.0 | 0.0 | 20.0 |
| 11 | 20.0 | 131.0 | 34.0 | 174.0 |
| 12 | 93.0 | 76.0 | 68.0 | 122.0 |
| 13 | 76.0 | 145.0 | 26.0 | 112.0 |
| 14 | 75.25 | 129.25 | 70.25 | 84.25 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.132 | 0.288 | 0.322 | 0.258 |
| 02 | 0.548 | 0.153 | 0.139 | 0.161 |
| 03 | 0.487 | 0.089 | 0.382 | 0.042 |
| 04 | 0.003 | 0.997 | 0.0 | 0.0 |
| 05 | 1.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.011 | 0.989 | 0.0 |
| 07 | 0.003 | 0.997 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 1.0 |
| 09 | 0.006 | 0.006 | 0.989 | 0.0 |
| 10 | 0.006 | 0.939 | 0.0 | 0.056 |
| 11 | 0.056 | 0.365 | 0.095 | 0.485 |
| 12 | 0.259 | 0.212 | 0.189 | 0.34 |
| 13 | 0.212 | 0.404 | 0.072 | 0.312 |
| 14 | 0.21 | 0.36 | 0.196 | 0.235 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.622 | 0.14 | 0.249 | 0.03 |
| 02 | 0.776 | -0.484 | -0.577 | -0.432 |
| 03 | 0.66 | -1.003 | 0.417 | -1.712 |
| 04 | -3.609 | 1.371 | -4.127 | -4.127 |
| 05 | 1.374 | -4.127 | -4.127 | -4.127 |
| 06 | -4.127 | -2.814 | 1.363 | -4.127 |
| 07 | -3.609 | 1.371 | -4.127 | -4.127 |
| 08 | -4.127 | -4.127 | -4.127 | 1.374 |
| 09 | -3.269 | -3.269 | 1.363 | -4.127 |
| 10 | -3.269 | 1.311 | -4.127 | -1.447 |
| 11 | -1.447 | 0.373 | -0.945 | 0.654 |
| 12 | 0.035 | -0.163 | -0.272 | 0.303 |
| 13 | -0.163 | 0.474 | -1.2 | 0.218 |
| 14 | -0.173 | 0.36 | -0.241 | -0.062 |
| P-value | Threshold |
|---|---|
| 0.001 | 2.55206 |
| 0.0005 | 3.96081 |
| 0.0001 | 6.36456 |