| Motif | ANDR.H14INVIVO.0.P.B |
| Gene (human) | AR (GeneCards) |
| Gene synonyms (human) | DHTR, NR3C4 |
| Gene (mouse) | Ar |
| Gene synonyms (mouse) | Nr3c4 |
| LOGO |  |
| LOGO (reverse complement) | 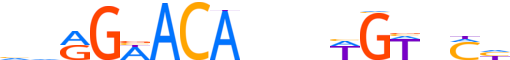 |
| Motif subtype | 0 |
| Quality | B |
| Motif | ANDR.H14INVIVO.0.P.B |
| Gene (human) | AR (GeneCards) |
| Gene synonyms (human) | DHTR, NR3C4 |
| Gene (mouse) | Ar |
| Gene synonyms (mouse) | Nr3c4 |
| LOGO |  |
| LOGO (reverse complement) | 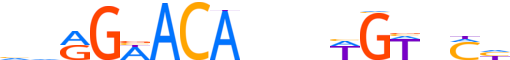 |
| Motif subtype | 0 |
| Quality | B |
| Motif length | 17 |
| Consensus | dKnWCWbnvTGTWCYbd |
| GC content | 47.22% |
| Information content (bits; total / per base) | 13.753 / 0.809 |
| Data sources | ChIP-Seq |
| Aligned words | 1000 |
| Previous names | ANDR.H12INVIVO.0.P.B |
| ChIP-Seq benchmarking | Num. of experiments (peaksets) | auROC, median | auROC, best | auPRC, median | auPRC, best | pseudo-auROC, median | pseudo-auROC, best | pseudo-log-auROC, median | pseudo-log-auROC, best | Centrality, median | Centrality, best |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | 237 (413) | 0.863 | 0.978 | 0.793 | 0.966 | 0.832 | 0.977 | 3.209 | 6.274 | 204.495 | 542.268 |
| Mouse | 20 (35) | 0.923 | 0.979 | 0.876 | 0.956 | 0.921 | 0.983 | 4.503 | 5.967 | 389.62 | 611.886 |
| HT-SELEX benchmarking | auROC10 | auPRC10 | auROC25 | auPRC25 | auROC50 | auPRC50 | |
|---|---|---|---|---|---|---|---|
| Non-Methyl HT-SELEX, 3 experiments | median | 0.694 | 0.708 | 0.586 | 0.606 | 0.539 | 0.559 |
| best | 0.999 | 0.998 | 0.992 | 0.99 | 0.918 | 0.917 | |
| rSNP benchmarking, ADASTRA | odds-ratio | -log-Fisher's P | Pearson r | Kendall tau |
|---|---|---|---|---|
| # | 1.154 | 1.832 | 0.032 | 0.005 |
| TF superclass | Zinc-coordinating DNA-binding domains {2} (TFClass) |
| TF class | Nuclear receptors with C4 zinc fingers {2.1} (TFClass) |
| TF family | Steroid hormone receptors {2.1.1} (TFClass) |
| TF subfamily | GR-like (NR3C) {2.1.1.1} (TFClass) |
| TFClass ID | TFClass: 2.1.1.1.4 |
| HGNC | HGNC:644 |
| MGI | MGI:88064 |
| EntrezGene (human) | GeneID:367 (SSTAR profile) |
| EntrezGene (mouse) | GeneID:11835 (SSTAR profile) |
| UniProt ID (human) | ANDR_HUMAN |
| UniProt ID (mouse) | ANDR_MOUSE |
| UniProt AC (human) | P10275 (TFClass) |
| UniProt AC (mouse) | P19091 (TFClass) |
| GRECO-DB-TF | yes |
| ChIP-Seq | 237 human, 20 mouse |
| HT-SELEX | 3 |
| Methyl-HT-SELEX | 0 |
| Genomic HT-SELEX | 0 overall: 0 Lysate, 0 IVT, 0 GFPIVT |
| SMiLE-Seq | 0 |
| PBM | 0 |
| PCM | ANDR.H14INVIVO.0.P.B.pcm |
| PWM | ANDR.H14INVIVO.0.P.B.pwm |
| PFM | ANDR.H14INVIVO.0.P.B.pfm |
| Threshold to P-value map | ANDR.H14INVIVO.0.P.B.thr |
| Motif in other formats | |
| JASPAR format | ANDR.H14INVIVO.0.P.B_jaspar_format.txt |
| MEME format | ANDR.H14INVIVO.0.P.B_meme_format.meme |
| Transfac format | ANDR.H14INVIVO.0.P.B_transfac_format.txt |
| Homer format | |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 416.0 | 18.0 | 334.0 | 232.0 |
| 02 | 120.0 | 18.0 | 732.0 | 130.0 |
| 03 | 273.0 | 250.0 | 248.0 | 229.0 |
| 04 | 776.0 | 51.0 | 66.0 | 107.0 |
| 05 | 4.0 | 948.0 | 26.0 | 22.0 |
| 06 | 669.0 | 30.0 | 79.0 | 222.0 |
| 07 | 121.0 | 266.0 | 340.0 | 273.0 |
| 08 | 309.0 | 201.0 | 228.0 | 262.0 |
| 09 | 264.0 | 340.0 | 281.0 | 115.0 |
| 10 | 21.0 | 19.0 | 3.0 | 957.0 |
| 11 | 2.0 | 2.0 | 995.0 | 1.0 |
| 12 | 8.0 | 3.0 | 0.0 | 989.0 |
| 13 | 126.0 | 91.0 | 94.0 | 689.0 |
| 14 | 7.0 | 979.0 | 0.0 | 14.0 |
| 15 | 4.0 | 511.0 | 2.0 | 483.0 |
| 16 | 162.0 | 252.0 | 183.0 | 403.0 |
| 17 | 167.0 | 128.0 | 310.0 | 395.0 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.416 | 0.018 | 0.334 | 0.232 |
| 02 | 0.12 | 0.018 | 0.732 | 0.13 |
| 03 | 0.273 | 0.25 | 0.248 | 0.229 |
| 04 | 0.776 | 0.051 | 0.066 | 0.107 |
| 05 | 0.004 | 0.948 | 0.026 | 0.022 |
| 06 | 0.669 | 0.03 | 0.079 | 0.222 |
| 07 | 0.121 | 0.266 | 0.34 | 0.273 |
| 08 | 0.309 | 0.201 | 0.228 | 0.262 |
| 09 | 0.264 | 0.34 | 0.281 | 0.115 |
| 10 | 0.021 | 0.019 | 0.003 | 0.957 |
| 11 | 0.002 | 0.002 | 0.995 | 0.001 |
| 12 | 0.008 | 0.003 | 0.0 | 0.989 |
| 13 | 0.126 | 0.091 | 0.094 | 0.689 |
| 14 | 0.007 | 0.979 | 0.0 | 0.014 |
| 15 | 0.004 | 0.511 | 0.002 | 0.483 |
| 16 | 0.162 | 0.252 | 0.183 | 0.403 |
| 17 | 0.167 | 0.128 | 0.31 | 0.395 |
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.506 | -2.546 | 0.288 | -0.074 |
| 02 | -0.727 | -2.546 | 1.07 | -0.648 |
| 03 | 0.087 | 0.0 | -0.008 | -0.087 |
| 04 | 1.128 | -1.563 | -1.313 | -0.84 |
| 05 | -3.783 | 1.328 | -2.206 | -2.362 |
| 06 | 0.98 | -2.071 | -1.137 | -0.118 |
| 07 | -0.718 | 0.062 | 0.306 | 0.087 |
| 08 | 0.211 | -0.216 | -0.091 | 0.047 |
| 09 | 0.054 | 0.306 | 0.116 | -0.769 |
| 10 | -2.405 | -2.497 | -3.975 | 1.337 |
| 11 | -4.213 | -4.213 | 1.376 | -4.525 |
| 12 | -3.253 | -3.975 | -4.982 | 1.37 |
| 13 | -0.678 | -0.999 | -0.967 | 1.009 |
| 14 | -3.362 | 1.36 | -4.982 | -2.773 |
| 15 | -3.783 | 0.711 | -4.213 | 0.655 |
| 16 | -0.43 | 0.008 | -0.309 | 0.475 |
| 17 | -0.4 | -0.663 | 0.214 | 0.455 |
| P-value | Threshold |
|---|---|
| 0.001 | 3.30296 |
| 0.0005 | 4.48506 |
| 0.0001 | 6.92891 |